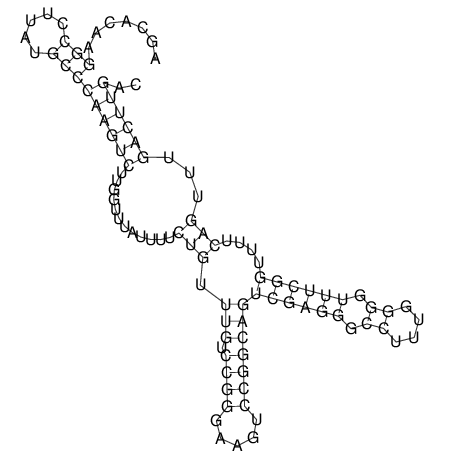
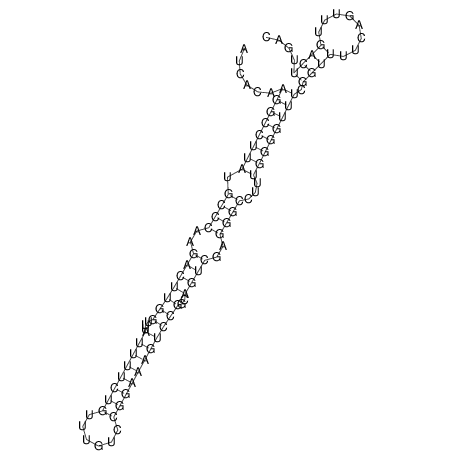
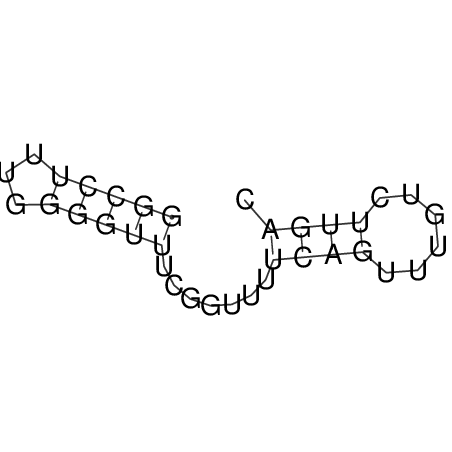
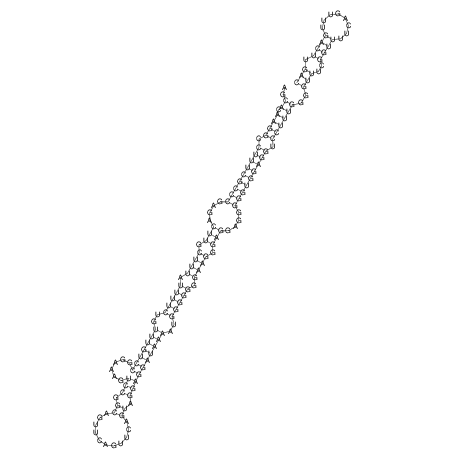
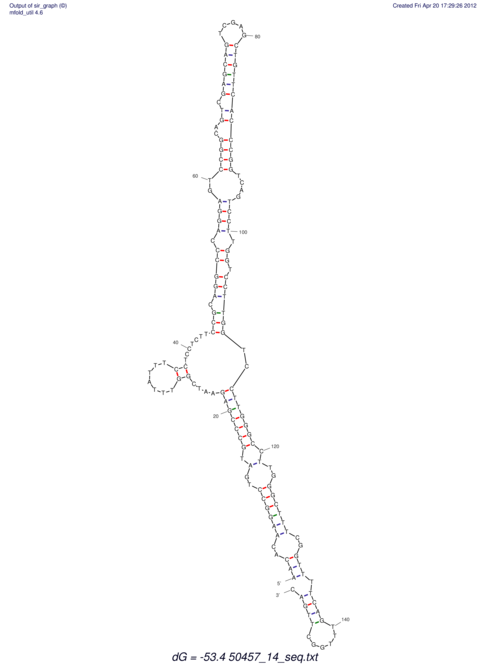
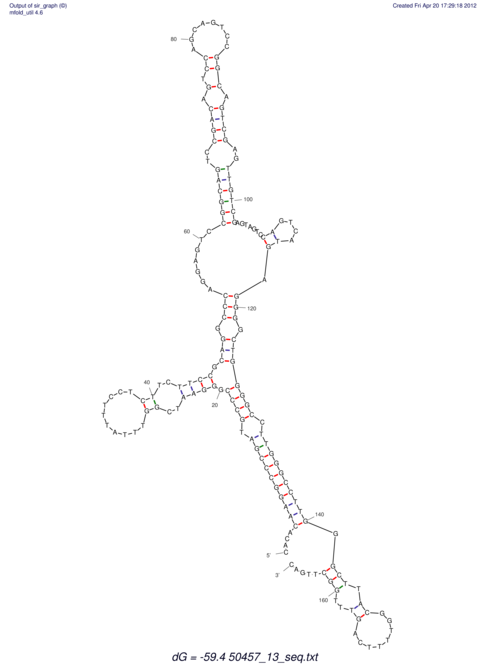
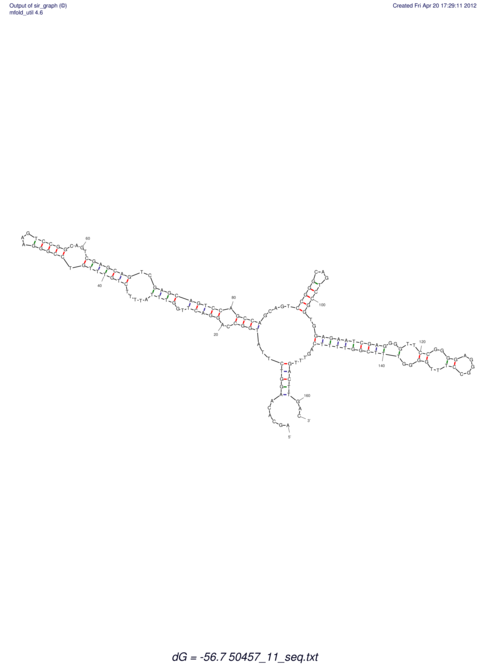
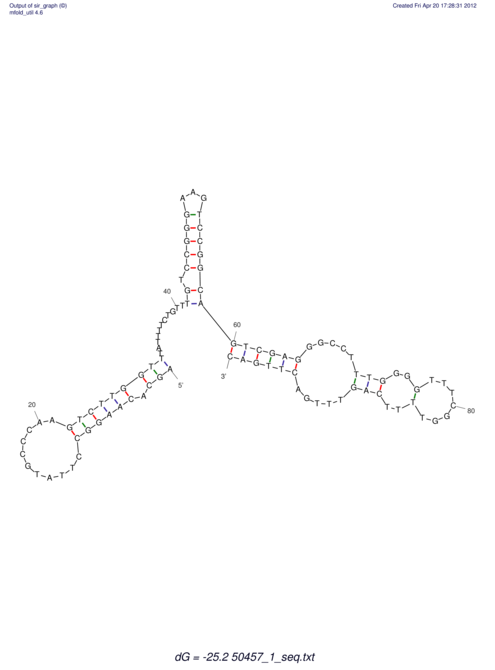
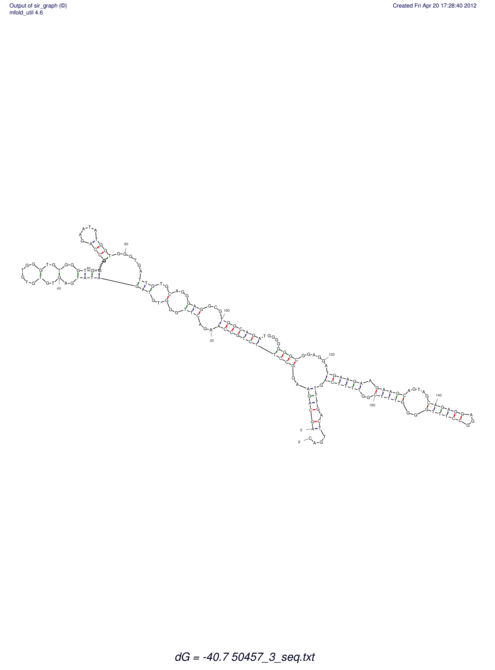
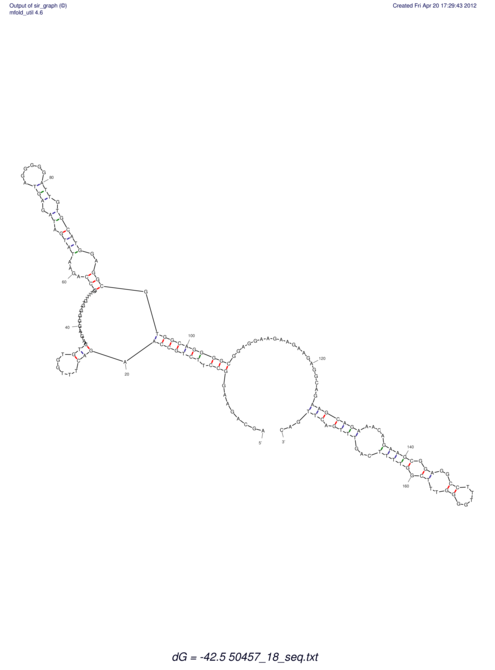
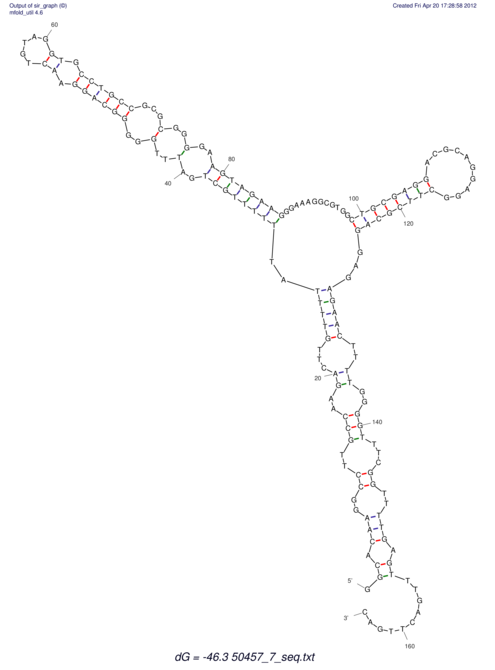
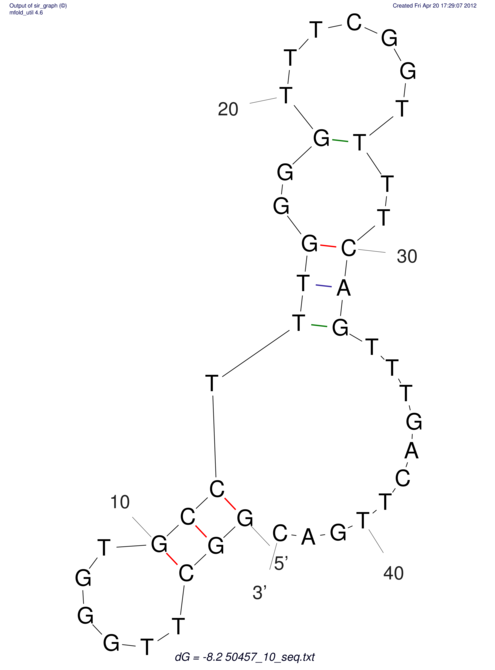
| dm3 |
chrX:463920-464051 + |
|
1 |
TGGAGTGACCAGC----------------------------AC--AACACAAGGCCTGCCGCCCAA-GAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCAAGCCCAGAAGTCC--------GGCAGTCGAG---------AA--------------------------------------------TCACGAGGGAC--TG------------------------------------GGACCTTGGGCTTACGGTTCTCAGTTTGGCTTGACATGCATCAGCATCAT |
| droSim1 |
chrX_random:776904-777034 + |
LASTZ |
4 |
CGGAGTGACCAGC----------------------------AC--ATCACAAGGCCTGGTGCCCAA-GAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCAAGCCCAGGAGTCC--------GGCAGTCGAG---------AA--------------------------------------------TCACGAGGGAC--TG-----------------------------------GGGCCTT--GGCTTACGGTTTTCATTTTGGCTTGACATGCATCAGCATCAT |
| droSec1 |
super_10:310837-310968 + |
LASTZ |
4 |
CGGAGTGACCAGC----------------------------AC--ATCACAAGGCCTGGTGCCCAA-GAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCAAGCCCAGGAGTCC--------GGCAGTCGAG---------AA--------------------------------------------TCACGAGGGAC--TG------------------------------------GGGCCTTGGGCTTACGGTTTTCATTTTGGCTTGACATGCATCAGCATCAT |
| droYak2 |
chrX:383437-383616 + |
LASTZ |
0 |
TGCAATGCCCACC----------------------------AC--AACACAAGGCCTGATGCCCGA-GAATCG-GTTTA----------------------------TTTCCTCCTCTTCCGC-------------------------------------------------------------------------------------------------------------------AGGCCCAGGAGTCC--------GGCAGTCGAGCAGTCGAGCTGTTCA--------------------CCCGGTCAG-----------TCCTT-GGTCC--TTGGTCCTT-----------------------------GGGCCTTGGGCTTTCGGTTTTCAGTTTGGCTTGACATGCATCAGCATCAT |
| droEre2 |
scaffold_4644:409107-409303 + |
LASTZ |
0 |
TGCAATGACCCTC----------------------------AC--CACACAAGGCCCGATGCCCGG-GAATCG-GTTTA----------------------------TTTCCTCTTCTTCCGC-------------------------------------------------------------------------------------------------------------------AGGCCCAGGAGTCC--------GGCAGTCCGACAGTCCAGCAGTCCG----GCAGTCGAGTTGTCGAGTAGTCCAG-----------TCATGAGGGGC--TGGGGCCTT-----------------------------GGGCCTTGGGCTTACGGTTTTCAGTTTGGCTTGACATGCATCAGCATCAT |
| droEug1 |
scf7180000409277:848596-848784 + |
LASTZ |
0 |
TGGAATGACC-------------------------------AC--AGCACAAGGTCTTATGGCCAG-GACTTG-GTTTA----------------------------TTTTCTGTTTGTCCGG------------------------------------------------------------------------------------------------------------------GA--------AGTCC--------GGCAGTCGAGCAGTCGAGCAGTCCAGCCAGCAGTC-------CGGGCAGTCCGGTGGAGAA----TC--GAGGGGTTC-C-GGG-----------------------------GAGGGCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droBia1 |
scf7180000301760:1515569-1515727 + |
LASTZ |
0 |
TGGAATGACCAGGACAGCACA-GCACAGCAGAGCACATAGCACATAGCACAAGGCCTTATGCCCAA-GTCTTG-GTTTA----------------------------TTTTCTGTTTGTCCGG------------------------------------------------------------------------------------------------------------------GA--------AGTCC--------GGCAGTCG----------------------------------------------------------------------------------------------------------AGGGCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droTak1 |
scf7180000415281:255352-255504 - |
LASTZ |
0 |
TGGAATGACCGGCACAGCACAAGCACA----AGCACA-AGCAC--ATCACAAGGCCTTATGCCCAA-GACTTG-GTTTA----------------------------TTTTCTGTTTGTCCGG------------------------------------------------------------------------------------------------------------------AA--------AGTCC--------GGCAGTCG----------------------------------------------------------------------------------------------------------AGGGCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droEle1 |
scf7180000491023:745050-745100 + |
LASTZ |
0 |
G---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCCTTTGGGGTTTCGGTTTTCAGTTTGTCTTGACATGCATCAGCATCAT |
| droRho1 |
scf7180000779514:240474-240645 + |
LASTZ |
0 |
TGGAATGACC-------------------------------AC--AGCACAAGGCCTTTCGCCCGA-GACTTG-CTTTA----------------------------TTTTCTGTTTGTCCGG------------------------------------------------------------------------------------------------------------------AA--------AGTCC--------GGCAGTTCAG---TTCAGTAGG----------------------AG-GATAAAATGGGGGGGAAGGGAGGAGGGGG--TG---------------------------------GAGGTCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droFic1 |
scf7180000453927:944869-945084 + |
LASTZ |
0 |
TGGAATGACC-------------------------------AC--AGCACAAGGCCTTCTGCCCAAAGACTGGGGTTTA----------------------------TTTTCTGTTTGTCGGG-----------------------------------------------------------------------------------------------------------------GAAA--------GTCCGGCAGTCCGGCAGTCGGG---------AGTCCAGTCG---ATC-------CGAG-AATCCAGTGGGAGG----TT--CGGGGGTTC-G-GGGCTTCGGGTTGGCAGAGAGTGTATGAAGTAGAGGGCCTTTGGGGTTTTGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droKik1 |
scf7180000302469:1859429-1859538 - |
LASTZ |
0 |
TGAAATGACC-------------------------------AC--AGCA----------------G-GTCCTG-CTTTT----------------------------CTTACTAGGGGG-CGA------------------------------------------------------------------------------------------------------------------AA--------TGGAA--------TGGAGGCG----------------------------------------------------------------------------------------------------------GTATCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droAna3 |
scaffold_12929:357992-358198 + |
LASTZ |
0 |
TGGAATGACC-------------------------------AC--AGCAGAAGGCCTTCTGCC-AA-GACTTGGGTGTAGATATGAGTGTGTGTGGGTGTGGGTGTGTTTT------G------GCCAGAATATGGT------GGGTGATTGTGCAGGGAGGCGTGGCAGATGGGGGGGCGG--------------------------AGGATGAAGAAGAAG------------CAGTAGCAGAGGA--------------------------------------------------------------------------------------------------------------------------------GGCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droBip1 |
scf7180000395176:32639-32844 - |
LASTZ |
0 |
TGGAATGACC-------------------------------AC--AGCAGAAGGCCTTCTGCC-AA-GACTTTGGTGTCAAT----------GTATGTGTGTGTGTGTTT------TG------GCCAGAATATGATAGAGTAGGGGGATTGTGCATGGAGGCGTGGCAG------GGGCGG--------------------------AGGAAGAAGAAGAGGCAGAAGCAGAAACAGAAGCGG-----------------------------------------------------------------------------------------------------------------------------------AGGCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| dp4 |
chrXL_group1e:196231-196421 + |
LASTZ |
0 |
TGGAATGACC-------------------------------AC--GGCACAAGGCCTT--G-CCAA-GACTTG-TTTTA----------------------------TTTTTTGCTGATTTGGGGGCAGGAACT-GT------AGGTGCC--------------TGCCGC------GCGGGGAAGTAGAAGGGAAAGGCGTGGCTGCGAG---GACGCAG-----------------GA--------GGCTT--------CGCAGGAG----------------------------------------------------------------------------------------------------------AGAACTTTTGGGGTTTCGGTTTTGAGTTTGACTTGACATGCATCAGCATCAT |
| droPer1 |
Unknown |
|
0 |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droWil1 |
scaffold_180777:2769923-2769983 + |
LASTZ |
0 |
CAG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGAT--TG-----------------------------------ACGCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droVir3 |
scaffold_12970:10710181-10710231 + |
LASTZ |
0 |
G---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droMoj3 |
scaffold_6473:10704148-10704209 - |
LASTZ |
0 |
GGGG---------------------------------------------------------------GGCTTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGTGCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droGri2 |
scaffold_14853:6069294-6069344 - |
LASTZ |
0 |
G---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |