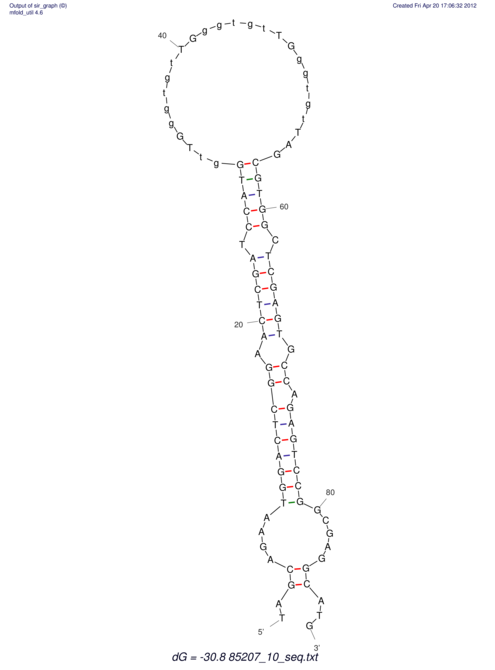
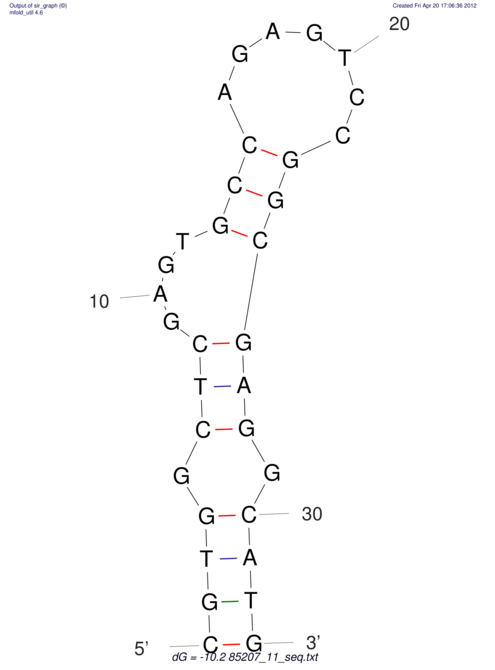
| dm3 |
chrX:1923021-1923142 - |
NA |
ATATAAGTGTTCGATTT---------------------GGCGCTTGGAATCGATACCCGAGCCATG---------------ATAGATTGAAGTCAAC-CC-AATC-------G-------ATCGCG--GTTCGAGTG-CTCGAGTCTTG-----TGCGCCGGCATGC--TTGAGAT---GTCCACTA |
| droSim1 |
Unknown |
NA |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droSec1 |
super_10:1708076-1708197 - |
1 |
ATATAAGTGTTCGATTT---------------------GGCGCTTGGAATCGACACCCGTGCCCTG---------------ATAGATTGAAGTCAAC-CC-AATC-------G-------ATCGCG--GTTCGGGTG-CTCGAGTCTTG-----TGCGCCGGCATGC--TTGAGAT---GTCCACTA |
| droYak2 |
chrX:6403257-6403382 + |
1 |
ACATAAAAGTTCAATTT---------------------GGCGCTTGGAATCGTAACTCGAGCCCTC---------------AAAGATTGAAGTCAAATGC-AATC-------G-------ATCCGGGGTGTCGAGTG-CTCGATTCTGG-----TGCGCCGGCATGC-TTTGAGAT---GTCCACTA |
| droEre2 |
scaffold_4644:1895276-1895400 - |
3 |
ATATAAGTGTTCGATTT---------------------GGCGCTTGGAATCGAACCTCGAGCCCTG---------------ACAGATTGAAGTCAAC-CCAAAAC-------G-------ATCCCGG-GGTCGAGTG-TTCGAGTCCGG-----TGCGCCGGCATGC-TTTGAGTT---GTCCACTA |
| droEug1 |
scf7180000409182:466737-466875 - |
1 |
ATATAAGTGTTGAATTT---------------------GGCGCTTGGAACCGAAACTCGATACGTGATCTATGATCCCTCAATAGGTTCAAGTTAAC-CA-AGGG-------G-------ATCTTT--GAAGCAGTG---CGAGTTCTG-----GGCGCCGGCATGT-TTTCAAATTATGTCCACTA |
| droBia1 |
scf7180000301760:4528531-4528646 - |
3 |
-TATAAGTGTTTAACTG---------------------GGCGCTTGAACCCGAAACTCGACCCCTG---------------AGA--ACGACCTCAAG-TC-AAGG-----------------------GATCAAGGG-ATCGAGTTTTG-----GGCGCCGGCATGT-TTTGAGATT-TGTACACTA |
| droTak1 |
scf7180000415211:15932-16040 + |
1 |
-TATAAGTGTTTAATTT---------------------GGCGCCTGCAACCGAAACTCGATCCCTG---------------ATATATT---------------------------------TCCCAGAAAATAAGTT-ATCGAGTTTTG-----AGCGCCGGCATGT-TTTGAGATGA--TTAACTA |
| droEle1 |
scf7180000491001:1438856-1438974 - |
2 |
-TATAGGTGTTTAATAA---------------------GGCGCTCGGAACCAAAACTCGATCCCTG---------------ATAGGATAAA--AAAC-CA-AAAA-------C-------ATTTTA--GATCCAGTG-TTCGAGTTTTG-----TGCGCCGAAAT------GAGATTGTGTCCACTA |
| droRho1 |
scf7180000777830:9566-9691 - |
2 |
-TATAAGTGTTTAATAT---------------------GGCGCTCGGAACCAAAACTCGATCCCTG---------------ATAGGATGAAGTAAAC-CG-AAAC-------C-------ATCTTG--TATCCAGTG-TTCGAGTTCTG-----TGCGCCGGCATGT-CTTGAGATTATGTCTACTA |
| droFic1 |
scf7180000454072:3036564-3036665 - |
4 |
-TACAGGTGTTTAAT-T---------------------GGCGCTTCGAAGCAAGACTCGAACTCGA---------------ACCCA--------------------------G-------ATCCTGG----ACAGGA-TTCGAGTTTTG-----TGCGCCGGCATGC-TC-CAGTA---TTCCAATA |
| droKik1 |
scf7180000302610:194767-194895 - |
3 |
ACATAACAATAATAATAACAATA----ACAATAGTGAGGATGCAGAGCTCTTAAGCCCGATCTATG---------------ATTGCCCTGTT-----------AA-------G-------ATCGTA--C---------ATCGAGTTTACTTACTAACCCCAAAATATGATTGTGAT---TGATACTA |
| droAna3 |
scaffold_12929:1917028-1917098 - |
1 |
ATATCCTTGACTTC------ATAAGGAACAAT--------------------------------------------------------------------------------G-------ATCATG------------T-----TAAGG-----TTCTCAGGCATGT-CTCGAGATTATATCGAATC |
| droBip1 |
scf7180000396543:377182-377261 - |
3 |
ATGTATGTATTTTAGGATCTATACCGAACAAA------------------------------------------------------------------------------------------------GATCAAGTTGTATGAGTT----------CTCAGGCATGT-CTCGAGTTTATATCGAGTA |
| dp4 |
chrXL_group1a:2597066-2597203 - |
2 |
ATATAAATACATACTGC---A----------TAGTGTAGCAGAATGGACTCGGAACTCGATCCATG---------------CATGCCACATGCCACATGC-CACATGTCGCAGAGGGGGCAGCGTG--GCTCGAGTG-CCAGAGTCC-------------GGCGAGG-CATGAGACTA---CTTACC |
| droPer1 |
super_28:1034311-1034357 - |
8 |
A-G-----------------------------------------------------------------------------------------------------------------------CGTG--GCTCGAGTG-CCAGAGTCC-------------GGCGAGG-CATGAGACTA---CTTACC |
| droWil1 |
Unknown |
NA |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
Unknown |
NA |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6473:2911987-2912050 + |
2 |
ATATATGTGTGTGTGTT---------------------TGTGTATTGTATTTAGATTTGAGCTATA---------------AAATGTTTAATAAAAC-TC-A------------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
NA |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |