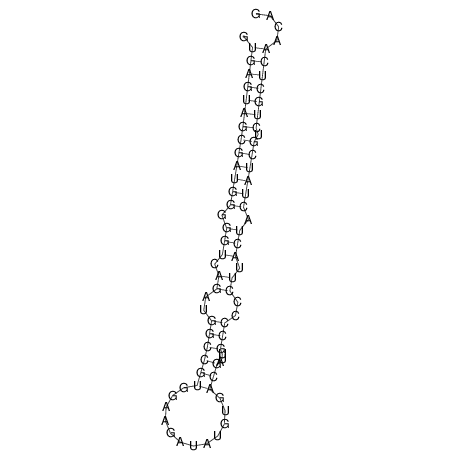
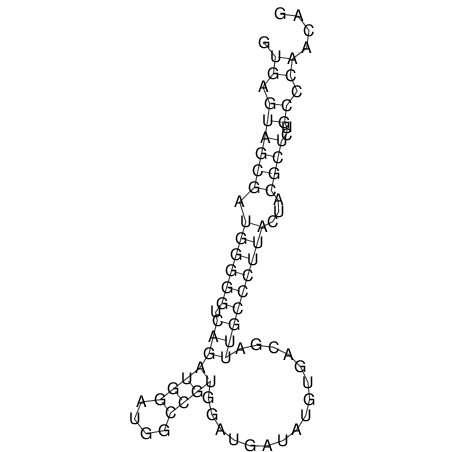
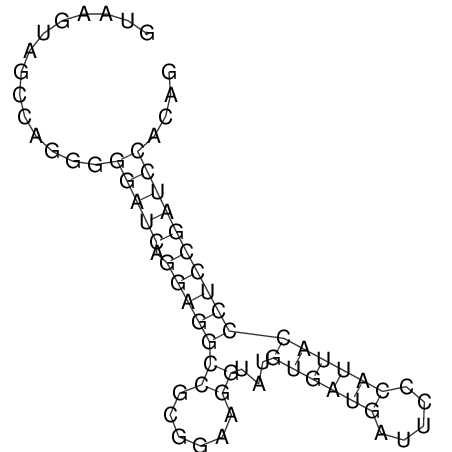
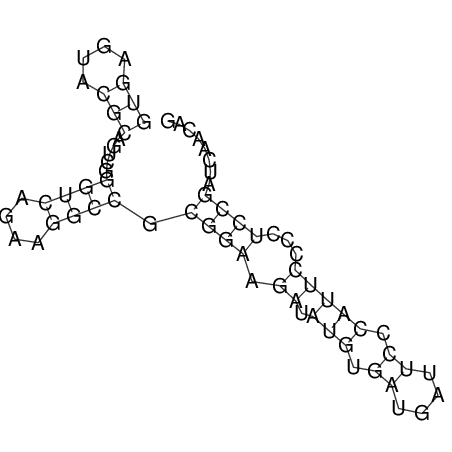
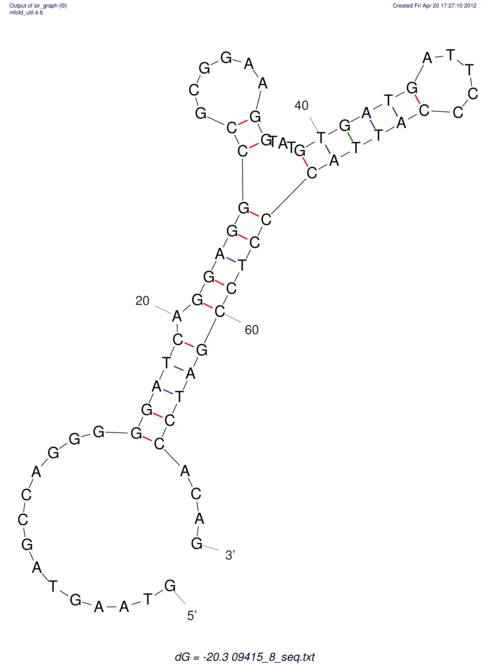
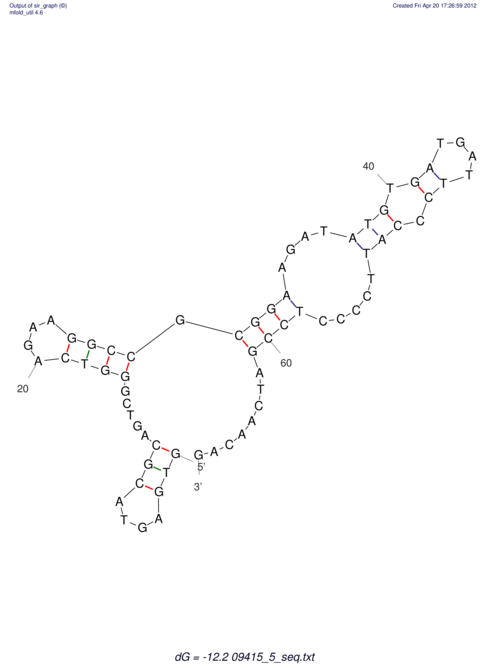
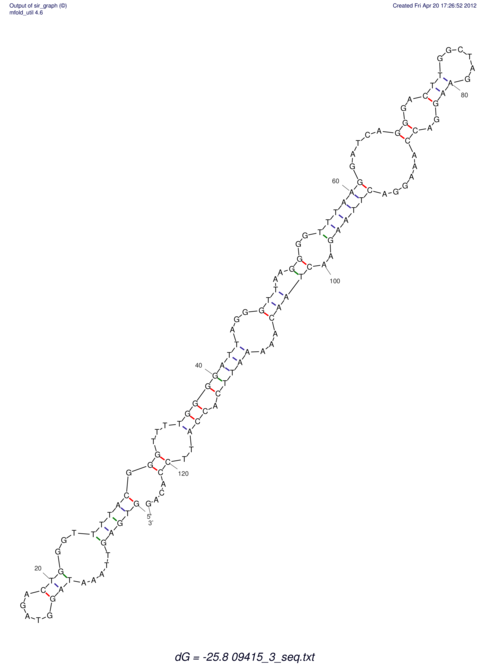
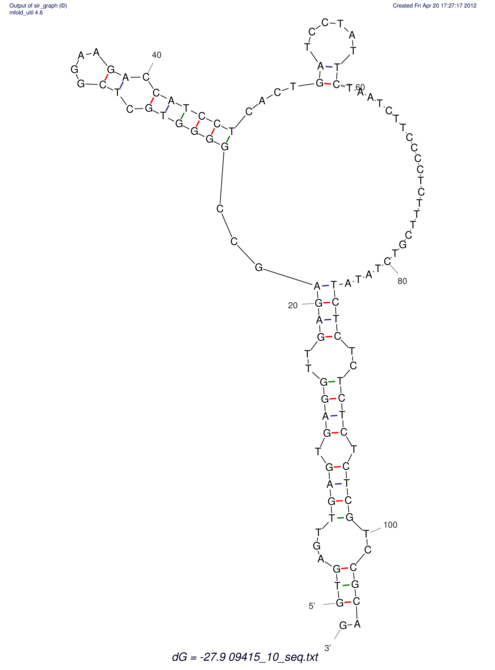
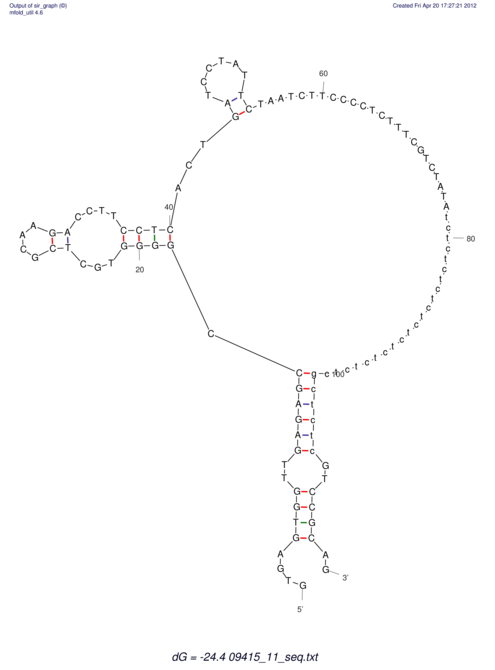
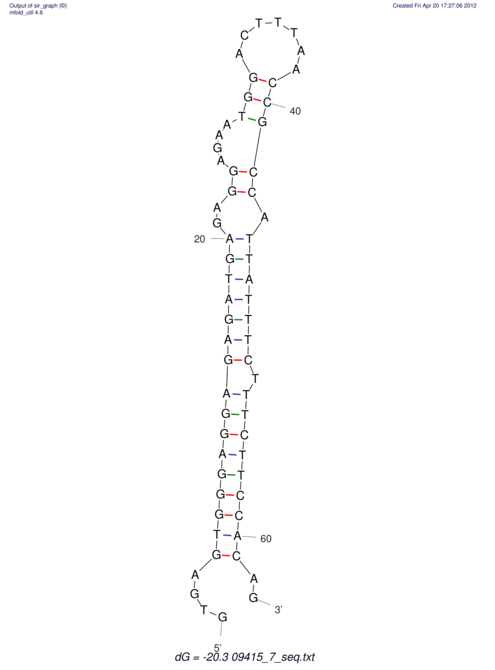
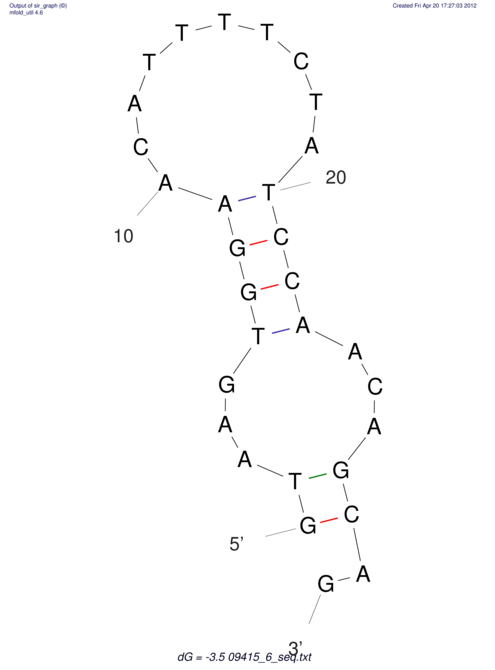
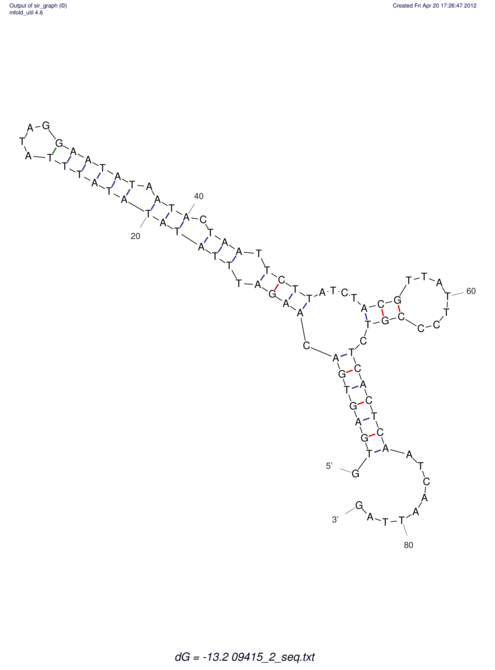
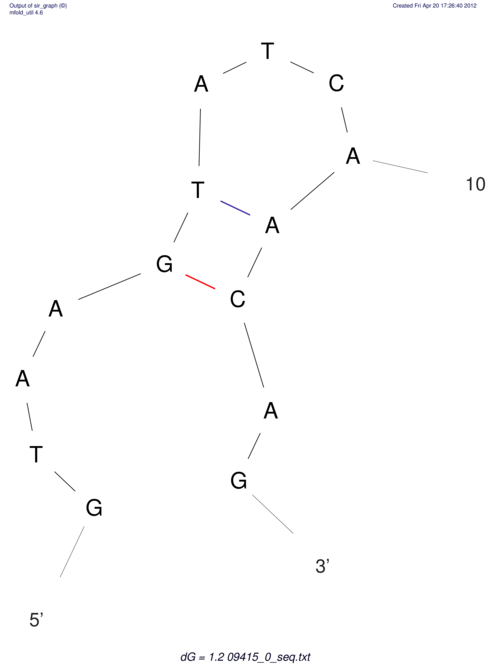
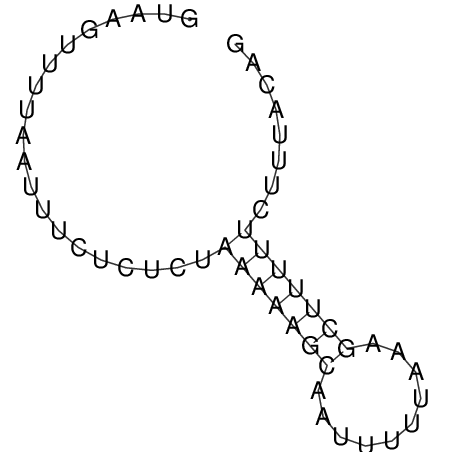
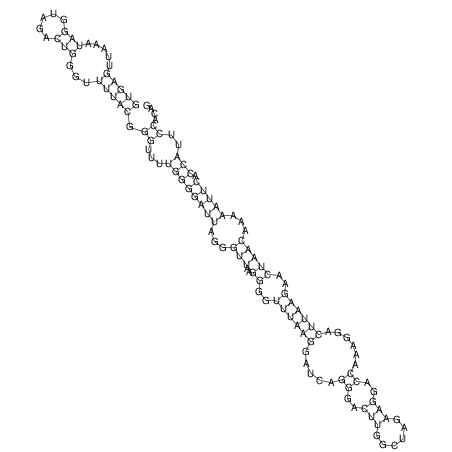
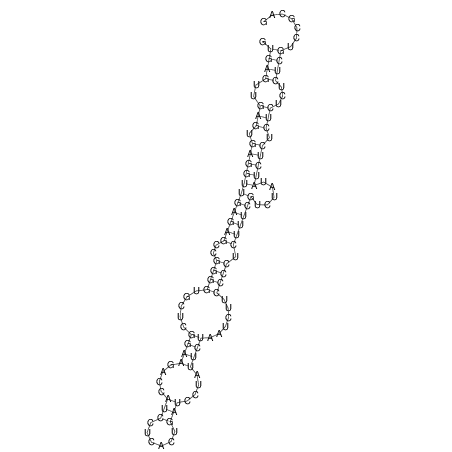
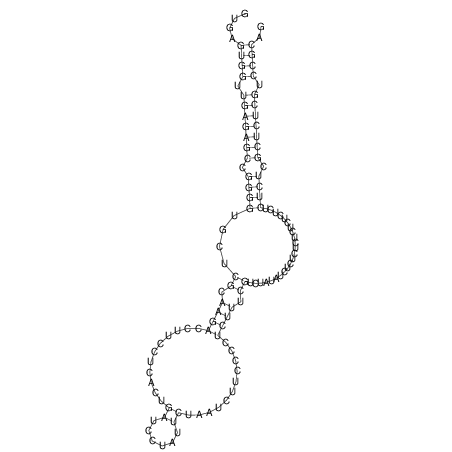
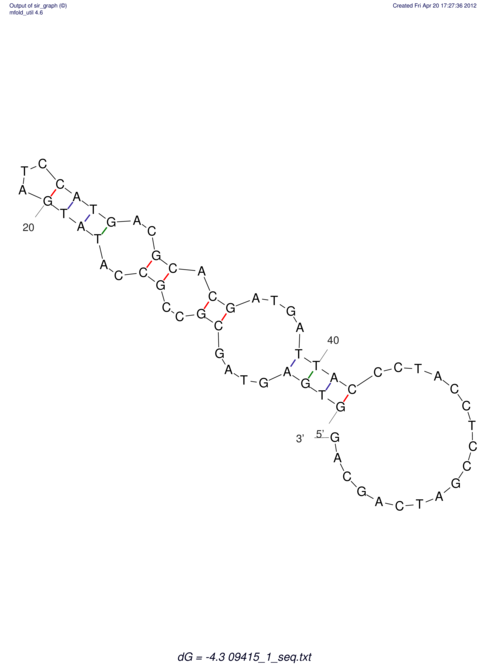
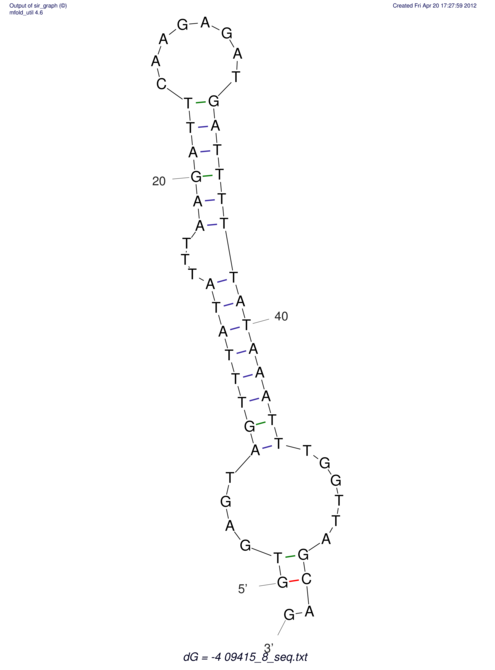
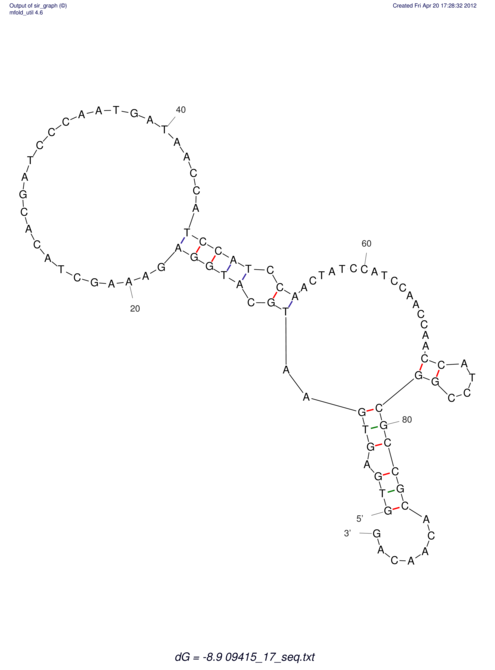
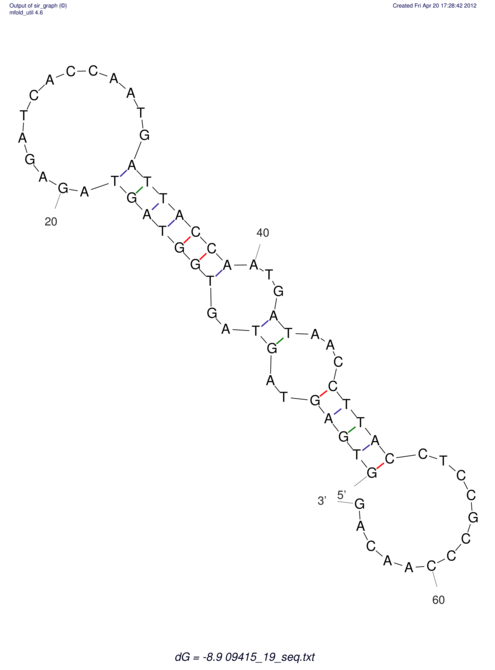
| GAAAACTCC---AGAGTGAGTGG------------------------------------------------TT--G---------------------AGAGCCGGGGT----GCTCGCA--AGA----------CCTTCCTCACTGATCCTATTCTAATCTTCCCCTCTTTCGTCTATAtctc--tctctctctctctctctcgctctc---GTCCGCAGGCAAGCGAACTATAC | Size | Hit Count | Total Norm | V111
Male body | V042
Embryo | V050
Head | V057
Head | M021
Embryo |
| ...................................................................................................................................................................................TCTC--TCTxGCGCTCTCTCTCT................................. | 20 | 2 | 1.50 | 0.00 | 0.00 | 0.00 | 0.00 | 1.50 |
| ....................................................................................................................................................................................CTG--TCTCTGTCTCTCTCT................................... | 18 | 3 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ......................................................................................................................................................................................C--TGTCTCTGTCTCTCTCT................................. | 18 | 3 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ..........................................................................................................................................................................................CTCTCTCTCTCTGTCGCGC.............................. | 19 | 2 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ......................................................................................................................................................................................C--TCGCTCTCTCTCTCTCTCTCTCT........................... | 24 | 2 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................................................................................................................................CTC--GCTCTCTCTCTCTCTCTCTCT............................. | 24 | 2 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ............................................................................................................................................................................................CTCTCTCTCTCTGTCGCGC............................ | 19 | 2 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ..........................................................................................................................................................................................CTCTCTCTCTCTCTGTCTCT............................. | 20 | 6 | 0.17 | 0.00 | 0.17 | 0.00 | 0.00 | 0.00 |
| ......................................................................................................................................................................................C--TCTCTCTCTCTGTCCCT................................. | 18 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| .............................................................................................................................................................................CATATxTCTC--TCTCTCTCTCTCT..................................... | 22 | 2 | 0.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 |
| .....................................................................................................................................................................................TC--TCTCCCTCTCTCTTTCTC................................ | 20 | 2 | 0.50 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 |
| .................................................................................................................................................................................TCTCTC--TCTTTCTCTCTCT..................................... | 19 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| .............................................................................................................................................................................................TCTCTCTCTTTCTCTCTCT........................... | 19 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ............................................................................................................................................................................................CTCTTTCTCTCTCTCTCT............................. | 18 | 8 | 0.13 | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 |
| ....................................................................................................................................................................................CTC--TCTCTCTCTCTGTCTCT................................. | 20 | 6 | 0.17 | 0.00 | 0.17 | 0.00 | 0.00 | 0.00 |
| .........................................................................................................................................................................................TCTCTCTCTCTCTCGCTG................................ | 18 | 5 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 |
| .........................................................................................................................................................................................TCTCTCTCTTTCTCTCTCT............................... | 19 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ..................................................................................................................................................................................CTCTC--TCTCTCTCTCTCTCT................................... | 20 | 5 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 |
| ....................................................................................................................................................................................CTC--TCGCTCGCTCTCTCTC.................................. | 19 | 2 | 0.50 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 |
| ..................................................................................................................................................................................CTCTC--TCTCTCTCTGTCTCT................................... | 20 | 6 | 0.17 | 0.00 | 0.17 | 0.00 | 0.00 | 0.00 |
| ....................................................................................................................................................................................CTC--TCTCTCTCTGTCCCT................................... | 18 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| ....................................................................................................................................................................................CTC--TCTCTCTCTCTCTCTCT................................. | 20 | 5 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 |
| .............................................................................................................................................................................................TCTCTCTCTCTCTCGCTG............................ | 18 | 5 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 |
| ......................................................................................................................................................................................C--GCTCTGTCTCTCTCTCTCGCT............................. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................................................................................................................................................TC--TCTCTCTCTCTCTGTCTCTCT............................. | 23 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| ......................................................................................................................................................................................C--TCTCTCTCTGTCACTCTC................................ | 19 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| ....................................................................................................................................................................................CTC--TCTCTCTGTCACTCTC.................................. | 19 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| ...........................................................................................................................................................................................TCTCTCTCTCTCTCTCGCTG............................ | 20 | 3 | 0.33 | 0.00 | 0.00 | 0.00 | 0.00 | 0.33 |
| ..............................................................................................................................................................................CTCTTTCTC--TCTCTCTCT......................................... | 18 | 8 | 0.13 | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 |
| ......................................................................................................................................................................................C--TCTCTCTCTCTCTGTCTCT............................... | 20 | 6 | 0.17 | 0.00 | 0.17 | 0.00 | 0.00 | 0.00 |
| .....................................................................................................................................................................................TC--TCTCTCTCTCTCGCTG.................................. | 18 | 5 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 |
| ...................................................................................................................................................................................TCTC--TCTCTCTCTCGCTG.................................... | 18 | 5 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 |
| ......................................................................................................................................................................................C--TCTCTxGCGCTCTCTCTC................................ | 18 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.25 | 0.00 |
| ..............................................................................................................................................................................................CTCTCTCTCTCGCGCACT........................... | 18 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................................................................................................................................CTC--TCTxGCGCTCTCTCTC.................................. | 18 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.25 | 0.00 |
| ...................................................................................................................................................................................TCTC--TCCCTCTCTCTTTCTC.................................. | 20 | 2 | 0.50 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 |
| ............................................................................................................................................................................................CTCTCTCTCTCTGTCCCT............................. | 18 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| ......................................................................................................................................................................................C--TCTTTCTCTCTCTCTCT................................. | 18 | 8 | 0.13 | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 |
| .....................................................................................................................................................................................TC--TCTCTCTTTCTCTCTCT................................. | 19 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ..........................................................................................................................................................................................CTCTTTCTCTCTCTCTCT............................... | 18 | 8 | 0.13 | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 |
| ................................................................................................................................................................................ATAGCTC--TCTCTCTCTCCCTCT................................... | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................................................................................................................................................TC--TCTCTCTCTCTCTCGCTG................................ | 20 | 3 | 0.33 | 0.00 | 0.00 | 0.00 | 0.00 | 0.33 |
| ......................................................................................................................................................................................C--TCTCGCTCGCTCTCTCTC................................ | 19 | 2 | 0.50 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 |
| ...................................................................................................................................................................................TCTC--TCTCTCTCTCTTTGTC.................................. | 20 | 3 | 0.33 | 0.00 | 0.00 | 0.33 | 0.00 | 0.00 |
| .........................................................................................................................................................................................TCTCTCTCTCTCTCTGTCTCTCT........................... | 23 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| ..............................................................................................................................................................................................CTCTTTCTCTCTCTCTCT........................... | 18 | 8 | 0.13 | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 |
| ..........................................................................................................................................................................................CTCTCTCTCTCTCTCTCTCT............................. | 20 | 5 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 |
| ...........................................................................................................................................................................................TCTCTCTCTTTCTCTCTCT............................. | 19 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ...................................................................................................................................................................................TCTC--TCCCTCTCTCTTTCTCT................................. | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ...................................................................................................................................................................................TCTC--TCTCTCTCTCTGTCTCTCT............................... | 23 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| ..................................................................................................................................................................................CTCTT--TCTCTCTCTCTCT..................................... | 18 | 8 | 0.13 | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 |
| ....................................................................................................................................................................................CTC--GTTCTCTCTCTCTCT................................... | 18 | 3 | 0.33 | 0.00 | 0.00 | 0.00 | 0.00 | 0.33 |
| ............................................................................................................................................................................................CTCTCTCTCTCTCTCTCTCT........................... | 20 | 5 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 |
| ...................................................................................................................................................................................TCTC--TCTCTTTCTCTCTCT................................... | 19 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ............................................................................................................................................................................................CTCTCTCTCTCTCTGTCTCT........................... | 20 | 6 | 0.17 | 0.00 | 0.17 | 0.00 | 0.00 | 0.00 |
| ...................................................................................................................................................................................TCTC--TCTCTCTCTCTCGCTG.................................. | 20 | 3 | 0.33 | 0.00 | 0.00 | 0.00 | 0.00 | 0.33 |
| ..............................................................................................................................................................................................CTCTCTCTCTGTCACTCTC---....................... | 19 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| .....................................................................................................................................................................................TC--TCTCTCTCTCTCTTTGTC................................ | 20 | 3 | 0.33 | 0.00 | 0.00 | 0.33 | 0.00 | 0.00 |
| ......................................................................................................................................................................................C--TCGTTCTCTCTCTCTCT................................. | 18 | 3 | 0.33 | 0.00 | 0.00 | 0.00 | 0.00 | 0.33 |
| ........................................................................................................................................................................................................ATCGCTCTC---GTACGCAGG.............. | 18 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............................................................................................................................................................................CATATxTC--TCTCTCTCTCTCTCT................................... | 22 | 2 | 0.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 |
| ....................................................................................................................................................................................CTC--TTTCTCTCTCTCTCT................................... | 18 | 8 | 0.13 | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 |
| .................................................................................................................................................................................TCTCTC--TCTCTCTCTGTCTCTCT................................. | 23 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.25 |
| ......................................................................................................................................................................................C--TCTCTCTCTCTCTCTCTCT............................... | 20 | 5 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 |
| ...................................................................................................................................................................................TCTC--TxACTCTCACTCTCTCTC................................ | 21 | 2 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 |