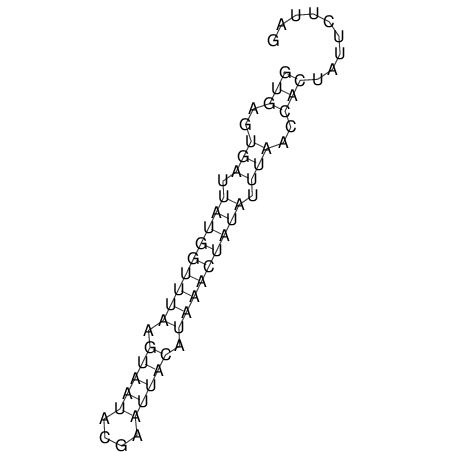
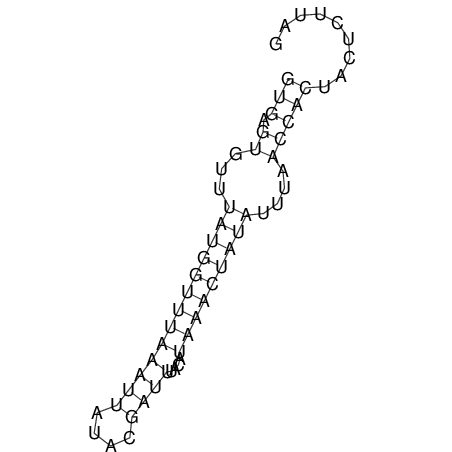
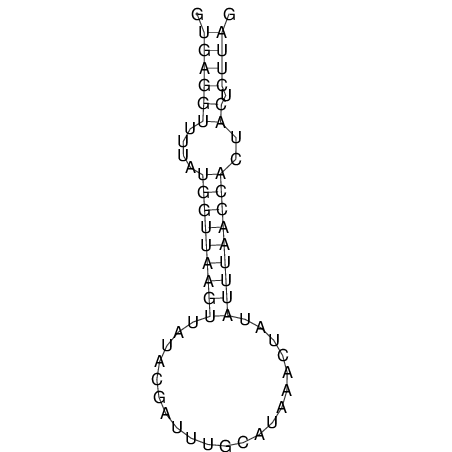
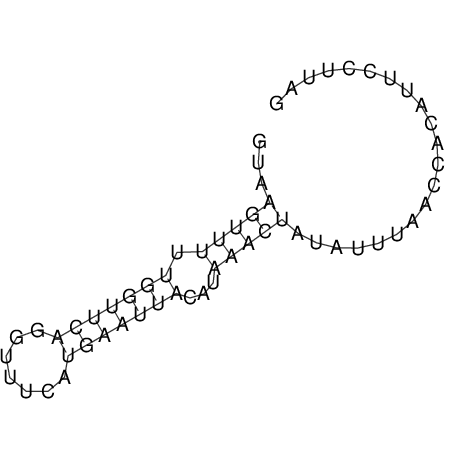
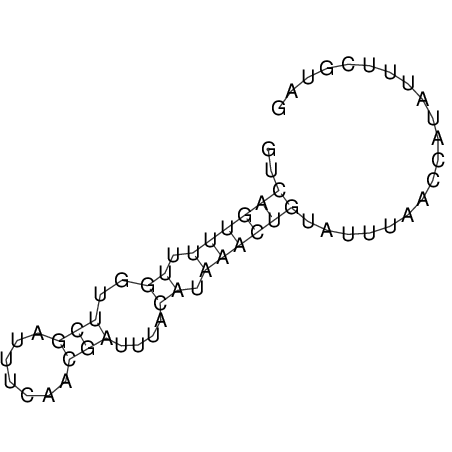
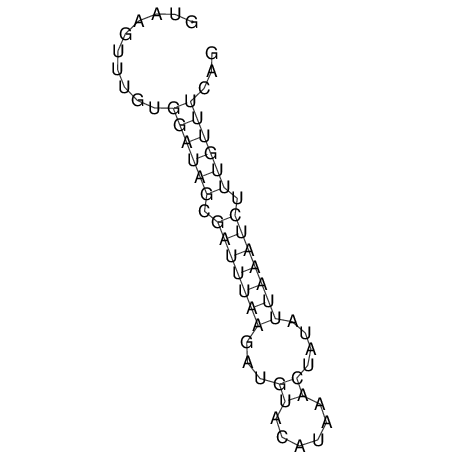
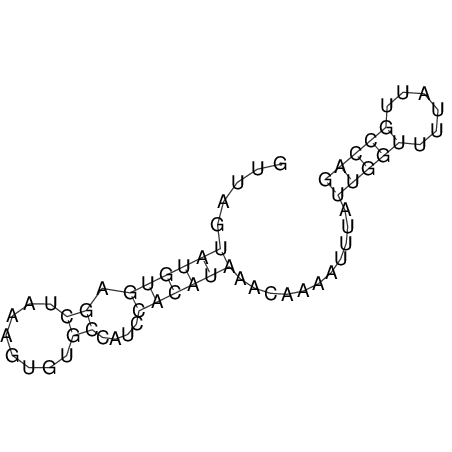
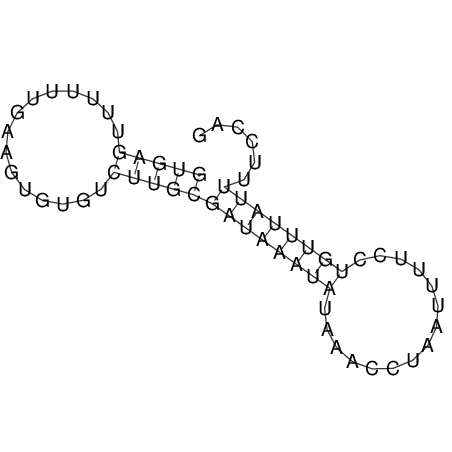
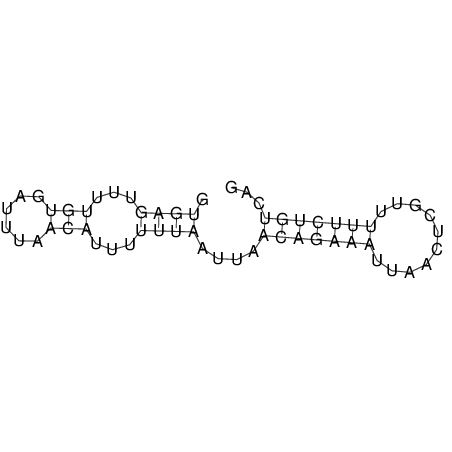
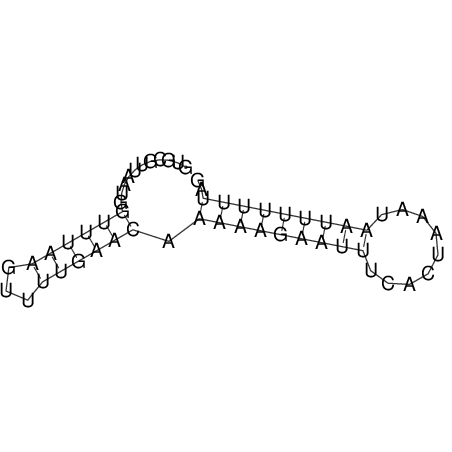
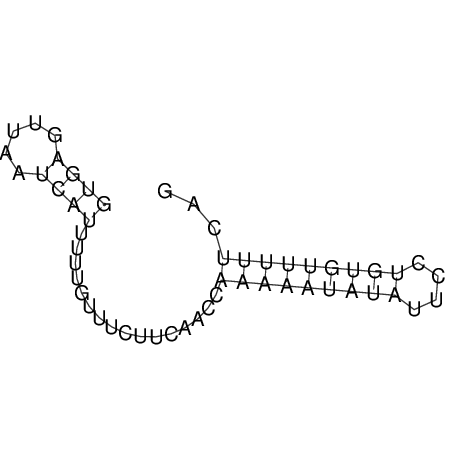
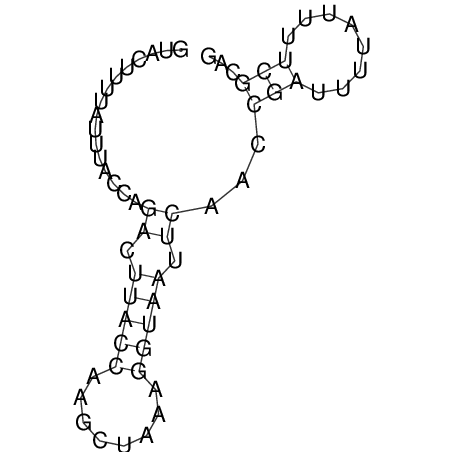
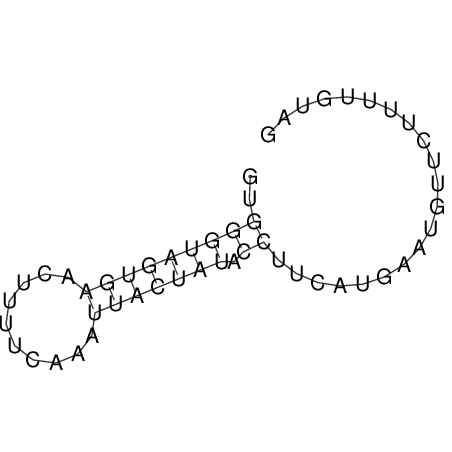
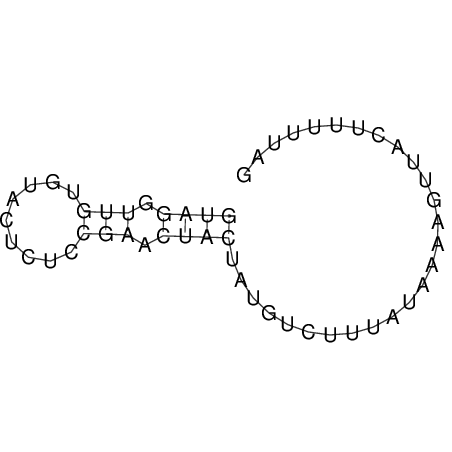
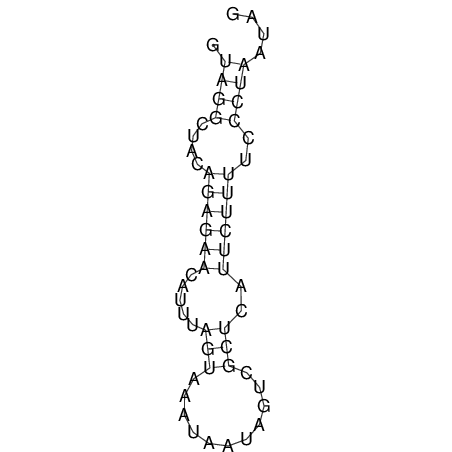
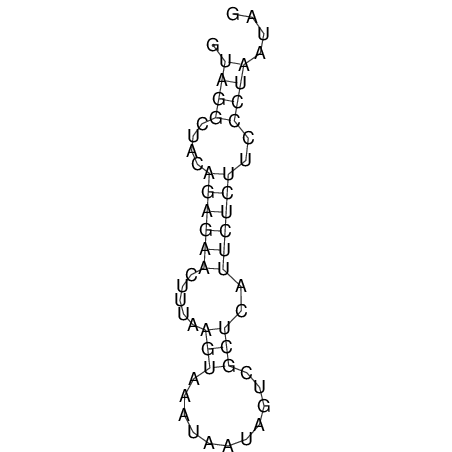
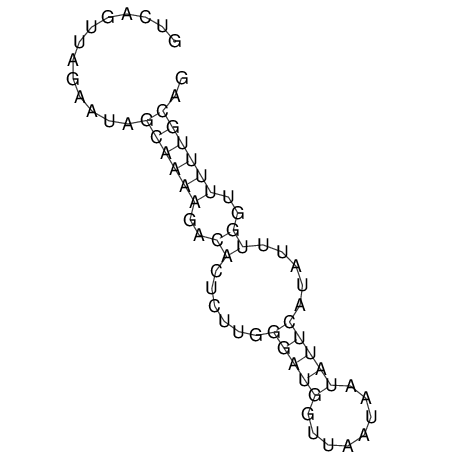
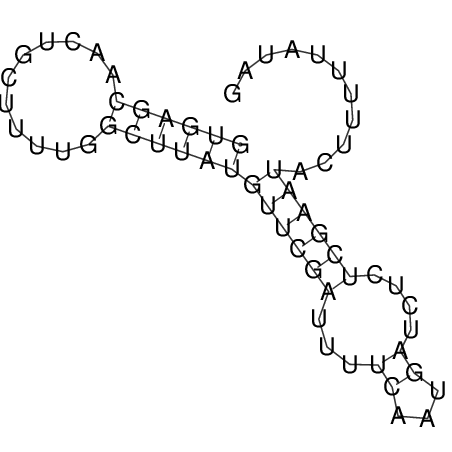
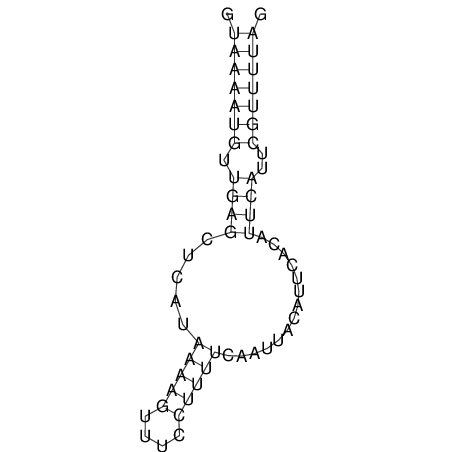
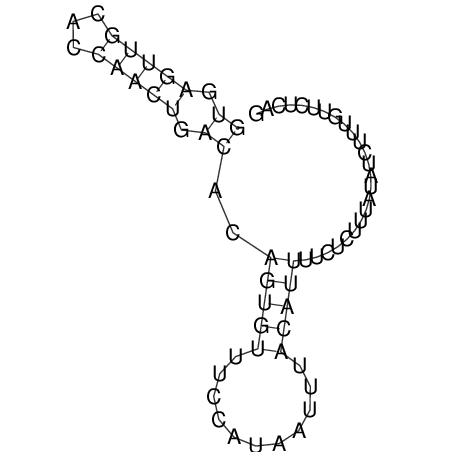
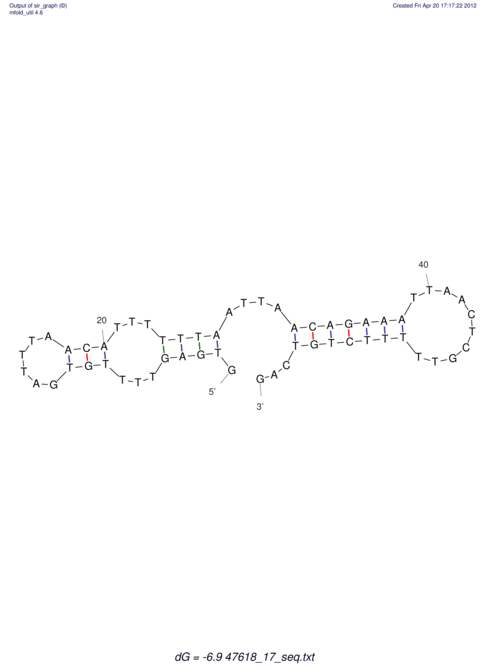
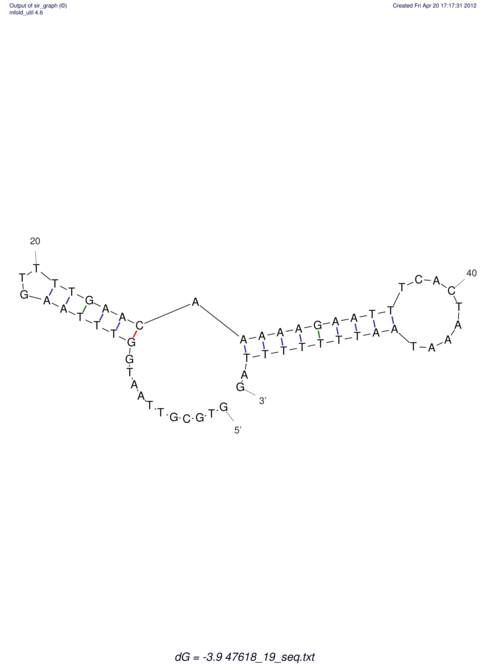
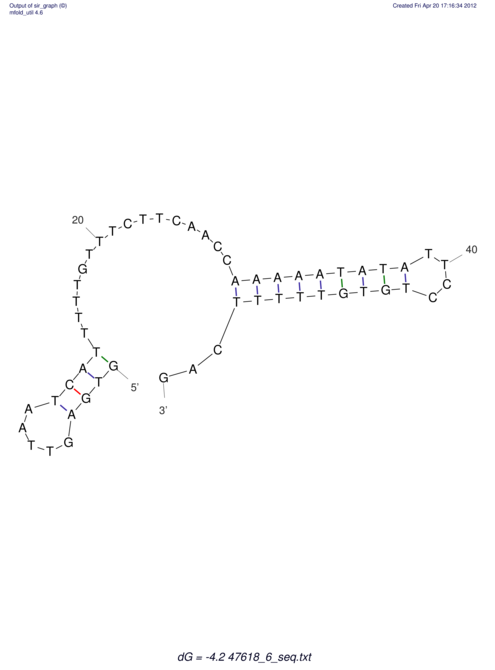
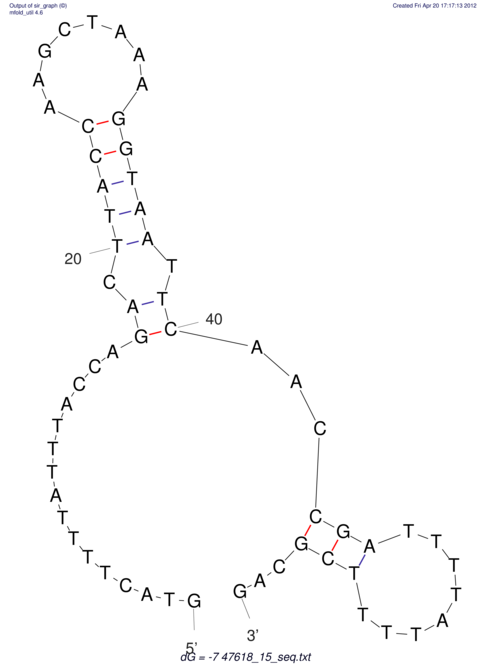
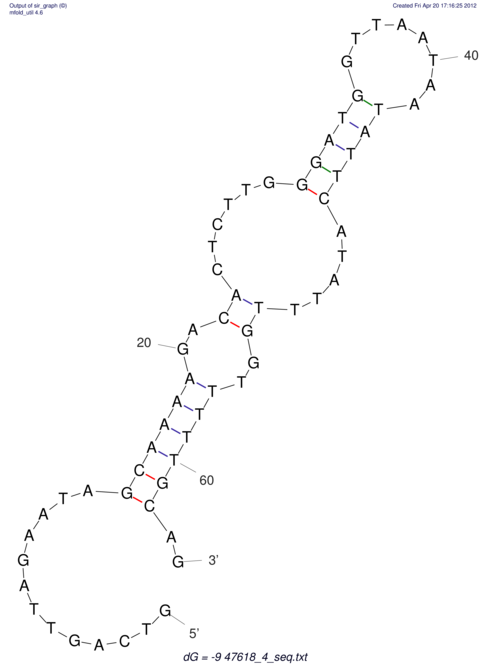
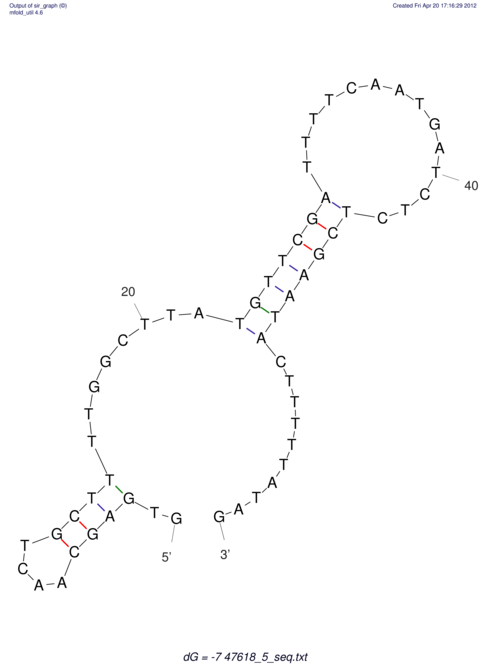
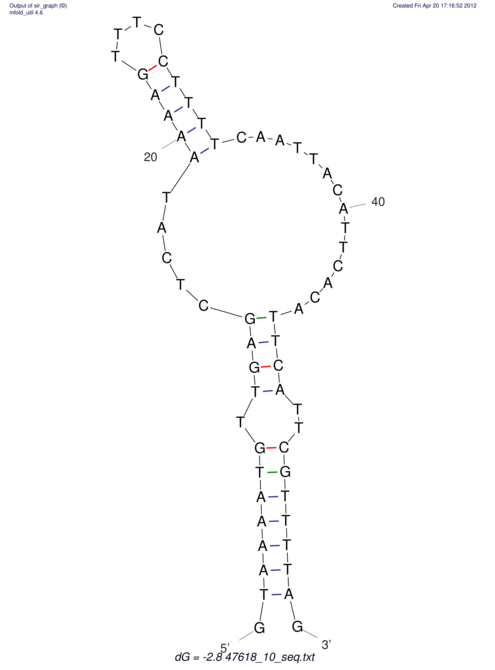
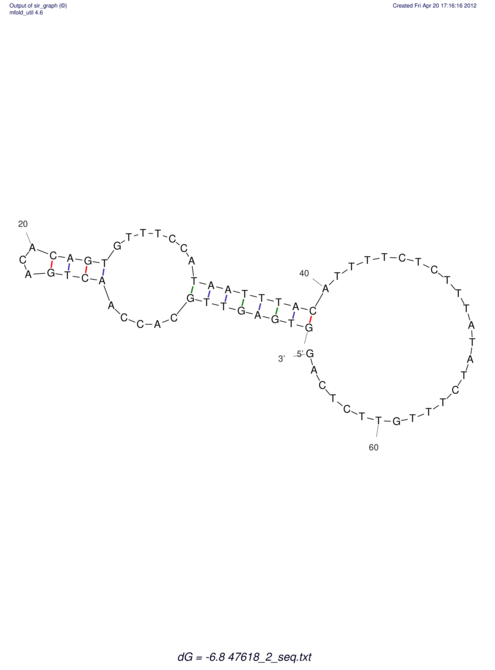
View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
dme-mir-3644 |
chr3R:11018019-11018079 - |
Known_mirbase |
|---|---|---|
| [View on miRBase] | [View on UCSC Genome Browser {Cornell Mirror}] | tailed_mirtron |
Legend:
mature strand at position 11018058-11018079| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr3R:11018004-11018094 - | 1 | TTATTGCTGCAGGAGGTGAGTGATTAT----GGTTTA--A--G--T----AATA---CGAATTACATAAACTAT----ATTT---AACCA--------CTA-TTCTTAGATACAAAACACTGCT | |
| droSim1 | chr3R:10448012-10448102 + | LASTZ | 4 | TTACTGCTGCAGGAGGTGAGTGTTTAT----GGTTTA--A--A--T----TATA---CGATTTACATAAACTAT----ATTT---AACCA--------CTA-CTCTTAGATCCAAAACACCGCT |
| droSec1 | super_0:10933739-10933828 + | LASTZ | 5 | TTACTGCTGCAGGAGGTGAGGTTTTAT----GGTT-A--A--G--T----TATA---CGATTTGCATAAACTAT----ATTT---AACCA--------CTA-CTCTTAGATCCAAAACACCGCT |
| droYak2 | chr3R:15416636-15416724 + | LASTZ | 5 | TTACTGCTTCAGGAGGTAAGTTTTTGG------TTCA--G--G--T----TTCA---TGAATTACATAAACTAT----ATTT---AACCA--------CAT-TCCTTAGATCCAAAATACCGCT |
| droEre2 | scaffold_4770:10539530-10539618 - | LASTZ | 5 | TTACTGCTTCAGGAGGTCAGTTTTTGG------TTCG--A--T--T----TCAA---CGATTTACATAAACTGT----ATTT---AACCA--------TAT-TTCGTAGATCCAAAACACCGCT |
| droEug1 | scf7180000409100:50607-50694 + | LASTZ | 5 | TTACTGCTGCAGGAGGTAAGTTTGTGG------ATAG--C--G--A----TTTA---AGATGTACATAAACTAT----ATTA---AATC---------TTT-GTTTCAGATCCAAAACACAGCT |
| droBia1 | scf7180000302402:3994917-3995007 - | LASTZ | 5 | TTGCTACTGCAGGAGGTTAGTATGTGA------GCTA--A--A--G----TGTG---CCATCCACATAAACAAA----ATTT---ATTGG------TTTTA-TTGCCAGATCCAAAACACTGCT |
| droTak1 | scf7180000415310:927278-927369 + | LASTZ | 5 | TTGCTTCTTCAGGAGGTGAGTTTTTTG-----AAGTG--T--G--T----CTTG---CGATAAATATAAACCTA----ATTT---TCCTG------TTTAT-TTTCCAGATCCAAAACACCGCT |
| droEle1 | scf7180000486474:959724-959811 + | LASTZ | 5 | TTGCTGCTGCAAGAGGTGAGTTTTG-T----GATTTA--A-CA--T----TTTT---TAATTAACAGAAA----------TT---AAC---T--CGTTTTT-CTGTCAGATCCAAAACACCGCT |
| droRho1 | scf7180000766350:39148-39233 - | LASTZ | 5 | TTACTGCTGCAAGAGGTGCGTTAATGG------TTTA--A--G--T----TT-----TGAACAAAAAGAA---------TTT---CACTA---A-ATAATT-TTTTTAGATCCAAAACACCGCT |
| droFic1 | scf7180000454104:344170-344253 + | LASTZ | 5 | TTGCTGCTGCAGGAGGTGAGTTAATCA------TTTT--T--G--T---TTCTT----CAACCA---AAAATAT----AT-----TCCTG--------TGT-TTTTCAGATACAAAACACAGCT |
| droKik1 | scf7180000301612:29528-29617 - | LASTZ | 5 | TTGCTGCTTCAAGAGGTACTTTTATTT------ACCA--G--A--C----TTAC---CAAGCTAAAGG----TA----ATTC---AACCG---ATTT-TATTTTCGCAGATCCAGAACACCGCT |
| droAna3 | scaffold_13340:6107797-6107878 - | LASTZ | 5 | TTGTTGCTGCAGGAGGTGGGTAGTG--------------A--A--C----TTTT---CAAATTACTATAC---------CTT------CATGAA-TGTTCT-TTTGTAGATTCAAAATACTGCG |
| droBip1 | scf7180000396708:2731560-2731641 + | LASTZ | 5 | TTACTGCTGCAGGAGGTAGGTTGTG--------------T--A--C----TCTC---CGAACTACTATGT---------CTT------TATAAA-AGTTAC-TTTTTAGATCCAAAACACTGCA |
| dp4 | chr2:21002920-21003004 - | LASTZ | 5 | CTGTTGCTGCAGGAGGTAGGCTACAGA-------G----AACA--T----TTA------------GTAAATAAT----AGTC---GCTCATT--CTTTTCC-CTAATAGATCCAAAACACTGCC |
| droPer1 | super_3:3754054-3754138 - | LASTZ | 5 | CTGTTGCTGCAGGAGGTAGGCTACAGA----G-------AAC------------------TTTAAGTAAATAAT----AGTC---GCTCATT--CTCTTCC-CTAATAGATCCAAAACACTGCC |
| droWil1 | scaffold_181130:7245340-7245433 + | LASTZ | 5 | TTGCTATTGCAAGAGGTCAGTTAGAAT----AGCA----A--AAGACACTCTTGGGATGGTTAATA------AT----ATT-------CATA--TTTGGTT-TTTGCAGATTCAAAATACTGCC |
| droVir3 | scaffold_13047:7744104-7744193 - | LASTZ | 5 | CTGCTGCTTCAAGAGGTGAGCAACTGCTTTTGGCTTA--T--G---------TT---CGATTTTCAATGA---------TCTCTCGAATA--------CTT-TTTATAGATACAAAACACAGCG |
| droMoj3 | scaffold_6540:3682572-3682661 - | LASTZ | 5 | TTGCTGCTGCAAGAGGTAAAATGTTGA----G-CTCATAA--AAGTTTCCTTTT-------------CAATTAC----ATT-------CACA--TTCATTC-GTTTTAGATACAAAATACGGCC |
| droGri2 | scaffold_14830:3558248-3558343 - | LASTZ | 5 | CTGCTGCTGCAGGAGGTGAGTTGCACCAACTGACACA--------------------GTGTTTCCATAATTTACATTTTCTC------TTTATA-TCTTTG-TTCTCAGATACAAAACACGGCT |
| Species | Alignment |
| dm3 | TTATTGCTGCAGGAGGTGAGTGATTAT----GGTTTA--A--G--T----AATA---CGAATTACATAAACTAT----ATTT---AACCA--------CTA-TTCTTAGATACAAAACACTGCT |
| droSim1 | TTACTGCTGCAGGAGGTGAGTGTTTAT----GGTTTA--A--A--T----TATA---CGATTTACATAAACTAT----ATTT---AACCA--------CTA-CTCTTAGATCCAAAACACCGCT |
| droSec1 | TTACTGCTGCAGGAGGTGAGGTTTTAT----GGTT-A--A--G--T----TATA---CGATTTGCATAAACTAT----ATTT---AACCA--------CTA-CTCTTAGATCCAAAACACCGCT |
| droYak2 | TTACTGCTTCAGGAGGTAAGTTTTTGG------TTCA--G--G--T----TTCA---TGAATTACATAAACTAT----ATTT---AACCA--------CAT-TCCTTAGATCCAAAATACCGCT |
| droEre2 | TTACTGCTTCAGGAGGTCAGTTTTTGG------TTCG--A--T--T----TCAA---CGATTTACATAAACTGT----ATTT---AACCA--------TAT-TTCGTAGATCCAAAACACCGCT |
| droEug1 | TTACTGCTGCAGGAGGTAAGTTTGTGG------ATAG--C--G--A----TTTA---AGATGTACATAAACTAT----ATTA---AATC---------TTT-GTTTCAGATCCAAAACACAGCT |
| droBia1 | TTGCTACTGCAGGAGGTTAGTATGTGA------GCTA--A--A--G----TGTG---CCATCCACATAAACAAA----ATTT---ATTGG------TTTTA-TTGCCAGATCCAAAACACTGCT |
| droTak1 | TTGCTTCTTCAGGAGGTGAGTTTTTTG-----AAGTG--T--G--T----CTTG---CGATAAATATAAACCTA----ATTT---TCCTG------TTTAT-TTTCCAGATCCAAAACACCGCT |
| droEle1 | TTGCTGCTGCAAGAGGTGAGTTTTG-T----GATTTA--A-CA--T----TTTT---TAATTAACAGAAA----------TT---AAC---T--CGTTTTT-CTGTCAGATCCAAAACACCGCT |
| droRho1 | TTACTGCTGCAAGAGGTGCGTTAATGG------TTTA--A--G--T----TT-----TGAACAAAAAGAA---------TTT---CACTA---A-ATAATT-TTTTTAGATCCAAAACACCGCT |
| droFic1 | TTGCTGCTGCAGGAGGTGAGTTAATCA------TTTT--T--G--T---TTCTT----CAACCA---AAAATAT----AT-----TCCTG--------TGT-TTTTCAGATACAAAACACAGCT |
| droKik1 | TTGCTGCTTCAAGAGGTACTTTTATTT------ACCA--G--A--C----TTAC---CAAGCTAAAGG----TA----ATTC---AACCG---ATTT-TATTTTCGCAGATCCAGAACACCGCT |
| droAna3 | TTGTTGCTGCAGGAGGTGGGTAGTG--------------A--A--C----TTTT---CAAATTACTATAC---------CTT------CATGAA-TGTTCT-TTTGTAGATTCAAAATACTGCG |
| droBip1 | TTACTGCTGCAGGAGGTAGGTTGTG--------------T--A--C----TCTC---CGAACTACTATGT---------CTT------TATAAA-AGTTAC-TTTTTAGATCCAAAACACTGCA |
| dp4 | CTGTTGCTGCAGGAGGTAGGCTACAGA-------G----AACA--T----TTA------------GTAAATAAT----AGTC---GCTCATT--CTTTTCC-CTAATAGATCCAAAACACTGCC |
| droPer1 | CTGTTGCTGCAGGAGGTAGGCTACAGA----G-------AAC------------------TTTAAGTAAATAAT----AGTC---GCTCATT--CTCTTCC-CTAATAGATCCAAAACACTGCC |
| droWil1 | TTGCTATTGCAAGAGGTCAGTTAGAAT----AGCA----A--AAGACACTCTTGGGATGGTTAATA------AT----ATT-------CATA--TTTGGTT-TTTGCAGATTCAAAATACTGCC |
| droVir3 | CTGCTGCTTCAAGAGGTGAGCAACTGCTTTTGGCTTA--T--G---------TT---CGATTTTCAATGA---------TCTCTCGAATA--------CTT-TTTATAGATACAAAACACAGCG |
| droMoj3 | TTGCTGCTGCAAGAGGTAAAATGTTGA----G-CTCATAA--AAGTTTCCTTTT-------------CAATTAC----ATT-------CACA--TTCATTC-GTTTTAGATACAAAATACGGCC |
| droGri2 | CTGCTGCTGCAGGAGGTGAGTTGCACCAACTGACACA--------------------GTGTTTCCATAATTTACATTTTCTC------TTTATA-TCTTTG-TTCTCAGATACAAAACACGGCT |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
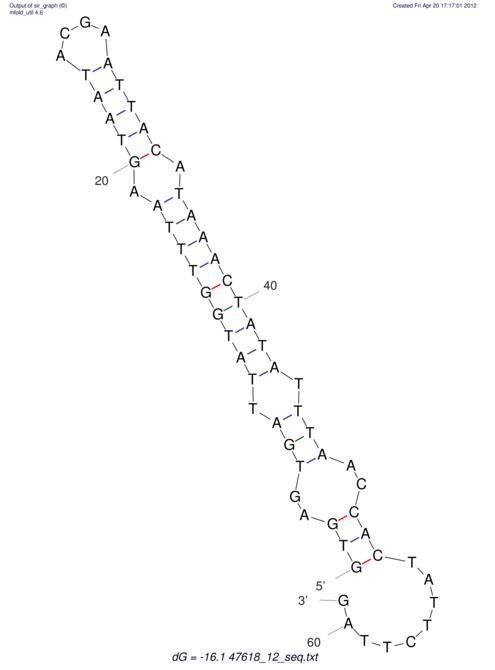
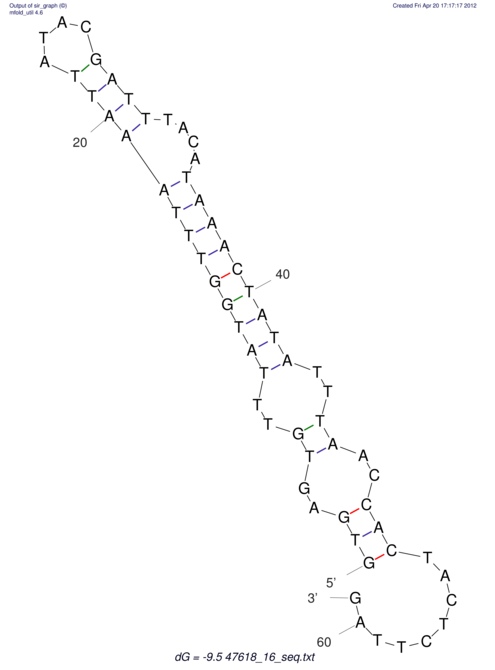
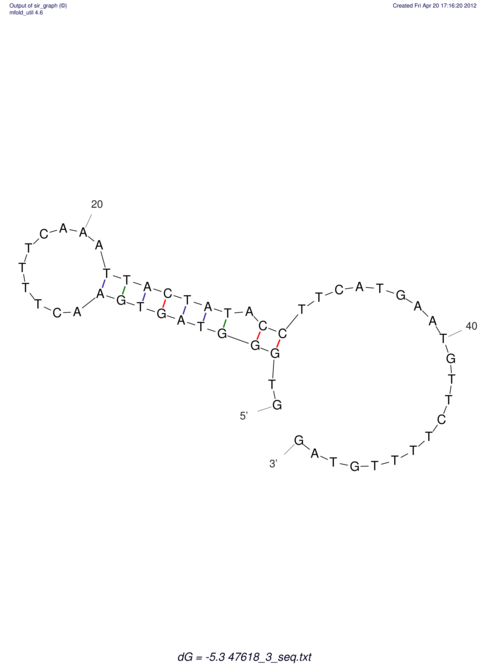
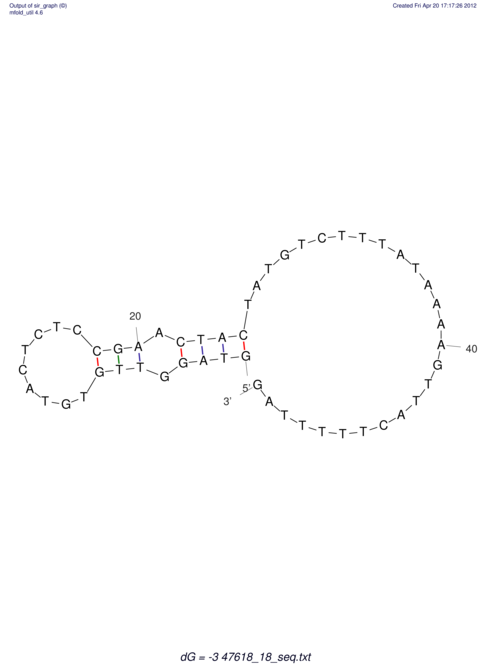
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
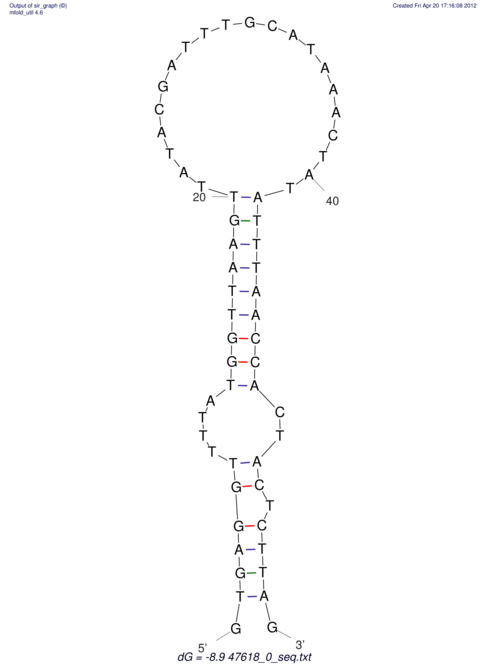
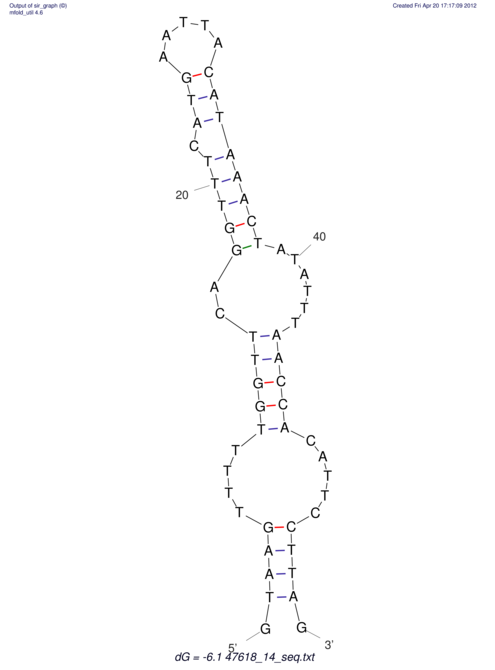
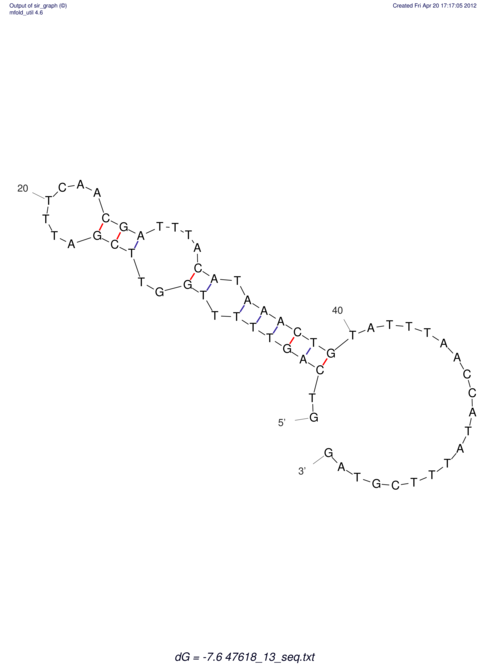
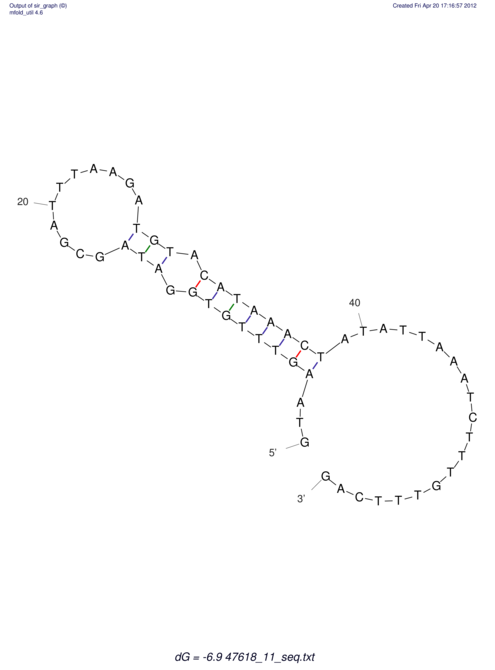
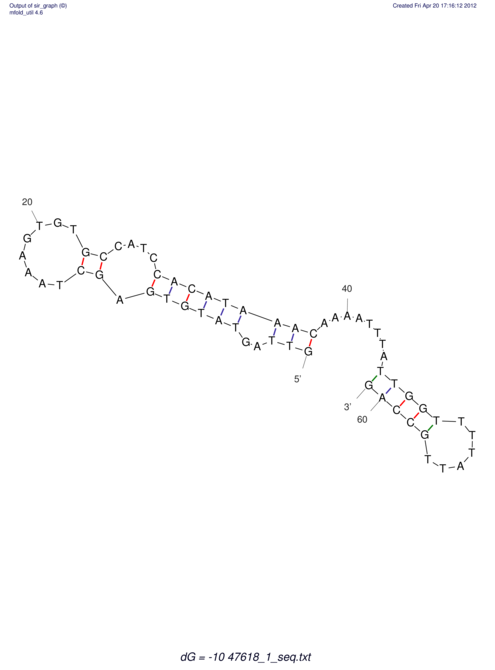
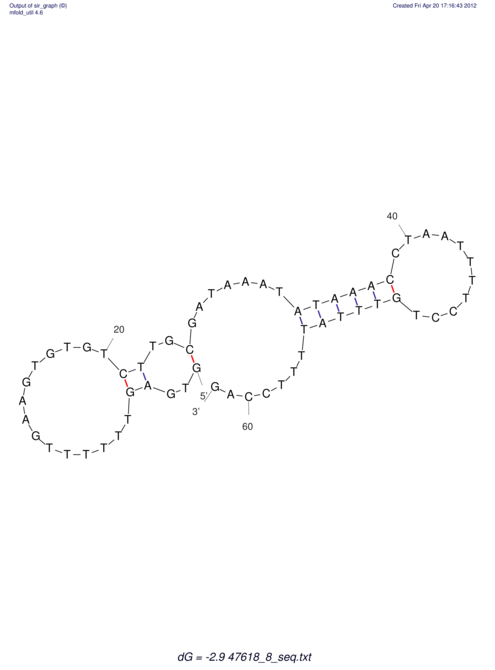
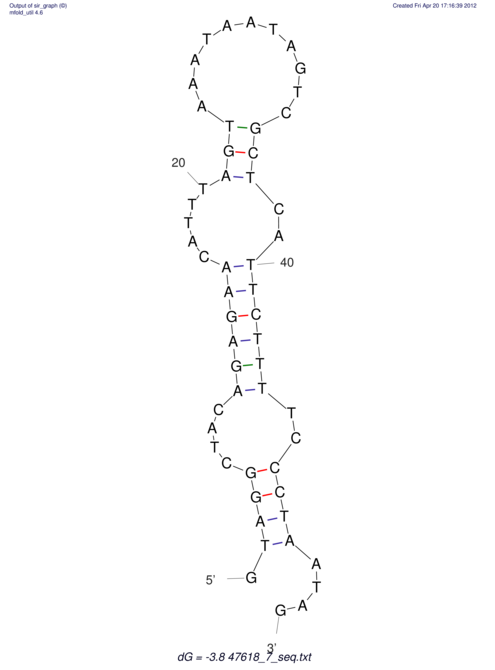
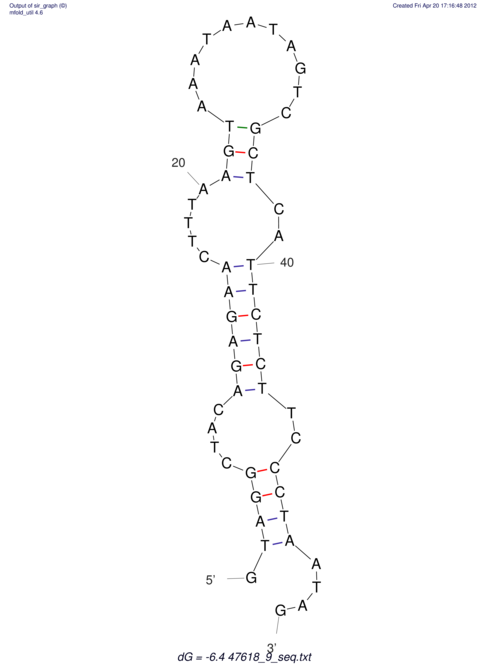
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:17:36 EDT