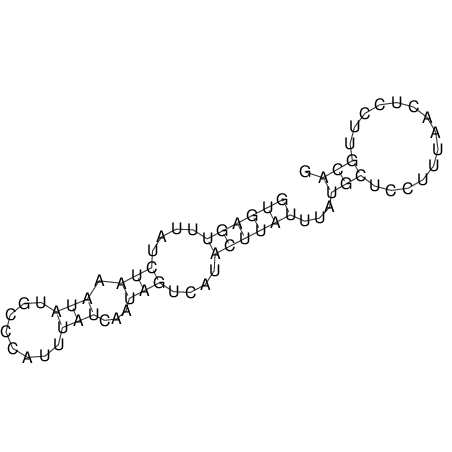
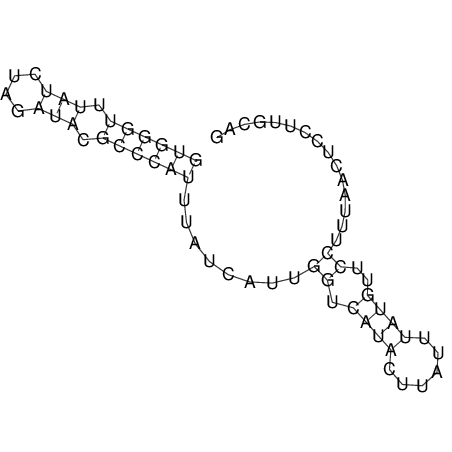

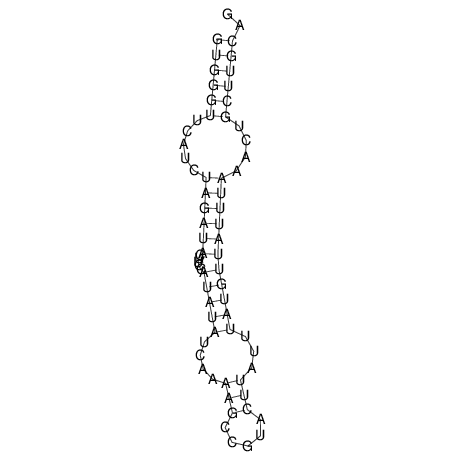
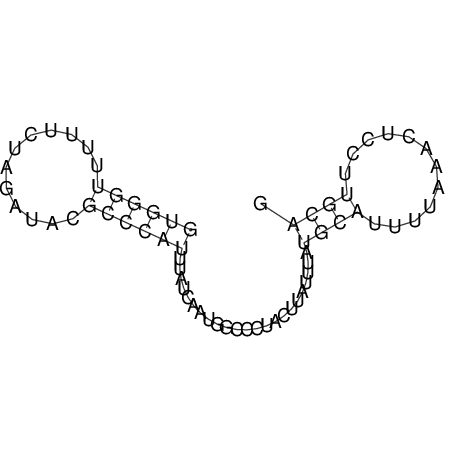

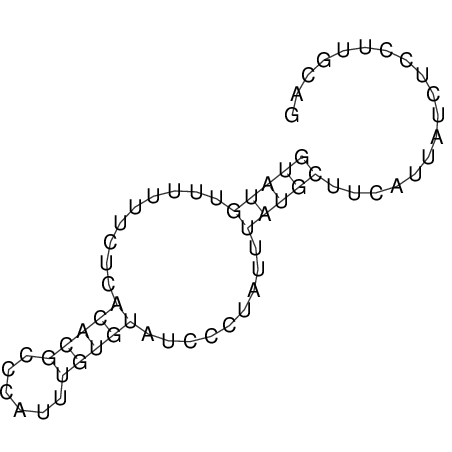
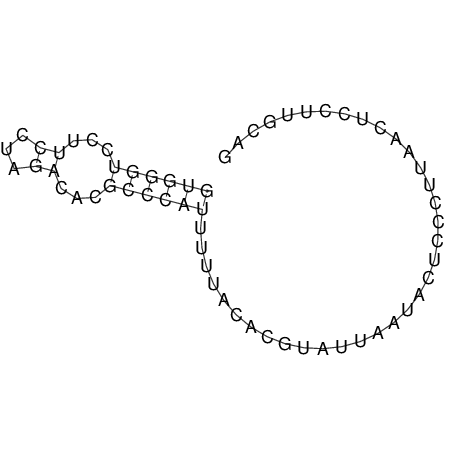
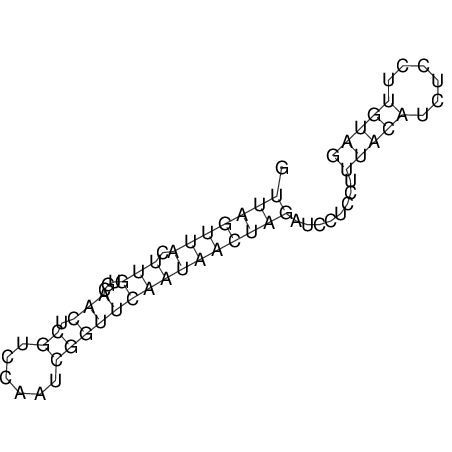
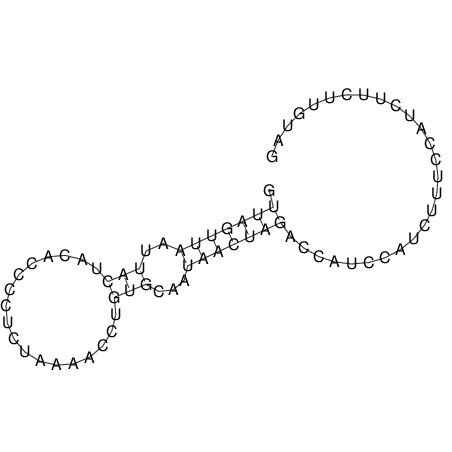







View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
dme-mir-3643 |
chr2R:13653898-13653965 - |
Known_mirbase |
|---|---|---|
| [View on miRBase] | [View on UCSC Genome Browser {Cornell Mirror}] | tailed_mirtron |
Legend:
mature strand at position 13653944-13653965| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr2R:13653883-13653980 - | 1 | GAATCCAAGCTGAACGTGAGTTTATC-TA--AATATG-CCCA-------------TTTATCAATAGTCATA--CTTATTTATG----------C--------T-----CCTTTAACTCCTTGCAGAGCTTCGAAAAAGAC | |
| droSim1 | chr2R:12393703-12393800 - | LASTZ | 5 | GAGTCCAAGCTGAACGTGGGTTTATC-TA--GATACG-CCCA-------------TTTATCATTGGTCATA--CTTATTTATG----------T--------T-----CCTTTAACTCCTTGCAGAGCTTCGAAAAAGAC |
| droSec1 | super_1:11152590-11152687 - | LASTZ | 5 | GAGTCCAAGCTGAACGTGGGTTTATC-TA--TTTACG-CCCA-------------TTTATCATTGGTCATA--CTTATTTATG----------T--------T-----CCTTTAACGCCTTGCAGAGCTTCGAAAAAGAC |
| droYak2 | chr2R:15527673-15527770 + | LASTZ | 5 | GAATCCAAGCTGAACGTGGGTTCATC-TA--GATACT-TCCA-------------TATATCAAAAGCCGTA--CTTATTTATG----------T--------T-----ATTTAAACTGCTTGCAGAGCTTCGAAAAAGAC |
| droEre2 | scaffold_4845:7883019-7883116 - | LASTZ | 5 | GAATCCAAGCTGAACGTGGGTTTTTC-TA--GATACG-CCCA-------------TTTATCAATGGCCCTA--CTTATTTATG----------C--------A-----TTTTAAACTCCTTGCAGAGCTTCGAAAAAGAC |
| droEug1 | scf7180000409672:1451984-1452077 + | LASTZ | 5 | GAATCCAAGCTGAACGTGAGTTCTTC-TA--TATACG-CCCA-------------CATGTCGGTACCTC------TTCTTATG----------C--------T-----TCCTTTACGTTTCGTAGAGTTTTGAAAAGGAC |
| droBia1 | scf7180000301754:2921050-2921150 + | LASTZ | 5 | GAATCCAAGCTGAACGTGCGTCTCTT-AAAACACACG-CCCA-------------TTTATCTGTATCCC------TACTTACGCTC-------G--------TC---CTTTTTTACTGATTGTAGAGCTTCGAGAAGGAC |
| droTak1 | scf7180000415313:36259-36358 + | LASTZ | 5 | GAATCCAAGCTGAACGTGGGTTCTGTCTA-ACATACG-CCGA-------------AATATATGTATCCC------TACTTATGCTA-----C-T--------C-----CTTTTGACTCCTTGCAGAGCTTCGAAAAGGAC |
| droEle1 | scf7180000491214:594780-594873 + | LASTZ | 5 | GAATCCAAGCTAAACGTGGGTTCTCC-CA--TATACG-CCCA-------------TTTGCCTGTACCC------CTATTTATG----------C--------T-----CCATTGACTTTTTGCAGAGCTTCGAAAAGGAC |
| droRho1 | scf7180000777956:135402-135495 + | LASTZ | 5 | GAATCCAAGCTGAACGTGGGTTCTCC-TA--TACACG-CCCA-------------TATGTCTGTACCCA------TGTTTATG----------C--------T-----CCTTTGACTCCTTGCAGAGCTTCGAAAAGGAC |
| droFic1 | scf7180000453948:1793989-1794080 + | LASTZ | 5 | GAATCCAAGCTGAACGTATGTTTTTT-CT--CACACG-CCCA-------------TTTGT--GTATC------CCTATTTATG----------C--------T-----TCATTATCTCCTTGCAGAGCTTCGAAAAGGAC |
| droKik1 | scf7180000302385:414803-414893 - | LASTZ | 5 | GAGTCCAAGCTGAACGTGGGTCCTTCCTA--GACACG-CCCA-------------TTTTT--------ACA--CGTATTAATA----------C--------T-----CCCTTAACTCCTTGCAGAGCTTCGAGAAGGAC |
| droAna3 | scaffold_13266:10531972-10532067 + | LASTZ | 5 | GAATCCAAGCTGAACGTTAGTTACTT-GTGCAACTCG-TCCA-------------ATCGG---TT-C--AATAACTA--GATC----------C--------TC---CTTTACATCTCCTTGTAGGGCTTCGAGAAGGAC |
| droBip1 | scf7180000396428:953655-953755 + | LASTZ | 5 | GAATCCAAGCTGAACGTTAGTTAATT-AC--TACACC-CCTC-------------TAAAACCTGTGCAATA--ACTA--GACC----------A--------TCCATCTTTCCATCTTCTTGTAGAGCTTCGAGAAGGAC |
| dp4 | chr3:16466931-16467019 - | LASTZ | 5 | GAATCCAAGCTGAACGTAAGTGCTTA-G---CAAATG-TTCACTGAACCTTGAAT------------------------CAT---------TGC--------C-----TTTGTTCTCGGTTCCAGAGCTTCGAAAAGGAC |
| droPer1 | super_4:295271-295359 + | LASTZ | 5 | GAATCCAAGCTGAACGTAAGTGCTTA-G---CAAATG-TTCACTGAACCTTGAAT------------------------CAT---------TGC--------C-----TTTGTTCTCGGTTCCAGAGCTTCGAAAAGGAC |
| droWil1 | scaffold_181141:4609865-4609965 + | LASTZ | 5 | GAATCAAAATTGAATGTAAGCTGA----------------------AAATTTAA-TTTATATTTTGT------TTTCTCCTAT---TTACA--TACCACGTGT-----CCTTCACTCAATTACAGAGCTTCGAAAAGGAT |
| droVir3 | scaffold_12875:16919908-16919994 - | LASTZ | 5 | GAATCCAAACTAAATGTGAGTATCTC-CT--AAT-CGCCTCA-------------TCCA-----------A--TTCATTTTTG----------A--------C-----CCTTTGCCTACTTTTAGAACTTTGAAAAAGAC |
| droMoj3 | scaffold_6496:11084070-11084155 + | LASTZ | 5 | GAATCTAAACTAAATGTAGGTATAGT-AA--TAGATG----AGTTTTTCTTTTAT------------------------TTT---------A-A--------C-----CTTTGGCCCGCTTGCAGAACTTTGAGAAGGAC |
| droGri2 | scaffold_15245:16459236-16459327 - | LASTZ | 5 | GAATCCAAACTAAATGTGAGATACCTCTA-AAACAAA-T-CA-------------TAAA----GATTTTGA--TGTATTGAGA--------------------------TTATGTACATTTGCAGAACTTTGAGAAAGAC |
| Species | Alignment |
| dm3 | GAATCCAAGCTGAACGTGAGTTTATC-TA--AATATG-CCCA-------------TTTATCAATAGTCATA--CTTATTTATG----------C--------T-----CCTTTAACTCCTTGCAGAGCTTCGAAAAAGAC |
| droSim1 | GAGTCCAAGCTGAACGTGGGTTTATC-TA--GATACG-CCCA-------------TTTATCATTGGTCATA--CTTATTTATG----------T--------T-----CCTTTAACTCCTTGCAGAGCTTCGAAAAAGAC |
| droSec1 | GAGTCCAAGCTGAACGTGGGTTTATC-TA--TTTACG-CCCA-------------TTTATCATTGGTCATA--CTTATTTATG----------T--------T-----CCTTTAACGCCTTGCAGAGCTTCGAAAAAGAC |
| droYak2 | GAATCCAAGCTGAACGTGGGTTCATC-TA--GATACT-TCCA-------------TATATCAAAAGCCGTA--CTTATTTATG----------T--------T-----ATTTAAACTGCTTGCAGAGCTTCGAAAAAGAC |
| droEre2 | GAATCCAAGCTGAACGTGGGTTTTTC-TA--GATACG-CCCA-------------TTTATCAATGGCCCTA--CTTATTTATG----------C--------A-----TTTTAAACTCCTTGCAGAGCTTCGAAAAAGAC |
| droEug1 | GAATCCAAGCTGAACGTGAGTTCTTC-TA--TATACG-CCCA-------------CATGTCGGTACCTC------TTCTTATG----------C--------T-----TCCTTTACGTTTCGTAGAGTTTTGAAAAGGAC |
| droBia1 | GAATCCAAGCTGAACGTGCGTCTCTT-AAAACACACG-CCCA-------------TTTATCTGTATCCC------TACTTACGCTC-------G--------TC---CTTTTTTACTGATTGTAGAGCTTCGAGAAGGAC |
| droTak1 | GAATCCAAGCTGAACGTGGGTTCTGTCTA-ACATACG-CCGA-------------AATATATGTATCCC------TACTTATGCTA-----C-T--------C-----CTTTTGACTCCTTGCAGAGCTTCGAAAAGGAC |
| droEle1 | GAATCCAAGCTAAACGTGGGTTCTCC-CA--TATACG-CCCA-------------TTTGCCTGTACCC------CTATTTATG----------C--------T-----CCATTGACTTTTTGCAGAGCTTCGAAAAGGAC |
| droRho1 | GAATCCAAGCTGAACGTGGGTTCTCC-TA--TACACG-CCCA-------------TATGTCTGTACCCA------TGTTTATG----------C--------T-----CCTTTGACTCCTTGCAGAGCTTCGAAAAGGAC |
| droFic1 | GAATCCAAGCTGAACGTATGTTTTTT-CT--CACACG-CCCA-------------TTTGT--GTATC------CCTATTTATG----------C--------T-----TCATTATCTCCTTGCAGAGCTTCGAAAAGGAC |
| droKik1 | GAGTCCAAGCTGAACGTGGGTCCTTCCTA--GACACG-CCCA-------------TTTTT--------ACA--CGTATTAATA----------C--------T-----CCCTTAACTCCTTGCAGAGCTTCGAGAAGGAC |
| droAna3 | GAATCCAAGCTGAACGTTAGTTACTT-GTGCAACTCG-TCCA-------------ATCGG---TT-C--AATAACTA--GATC----------C--------TC---CTTTACATCTCCTTGTAGGGCTTCGAGAAGGAC |
| droBip1 | GAATCCAAGCTGAACGTTAGTTAATT-AC--TACACC-CCTC-------------TAAAACCTGTGCAATA--ACTA--GACC----------A--------TCCATCTTTCCATCTTCTTGTAGAGCTTCGAGAAGGAC |
| dp4 | GAATCCAAGCTGAACGTAAGTGCTTA-G---CAAATG-TTCACTGAACCTTGAAT------------------------CAT---------TGC--------C-----TTTGTTCTCGGTTCCAGAGCTTCGAAAAGGAC |
| droPer1 | GAATCCAAGCTGAACGTAAGTGCTTA-G---CAAATG-TTCACTGAACCTTGAAT------------------------CAT---------TGC--------C-----TTTGTTCTCGGTTCCAGAGCTTCGAAAAGGAC |
| droWil1 | GAATCAAAATTGAATGTAAGCTGA----------------------AAATTTAA-TTTATATTTTGT------TTTCTCCTAT---TTACA--TACCACGTGT-----CCTTCACTCAATTACAGAGCTTCGAAAAGGAT |
| droVir3 | GAATCCAAACTAAATGTGAGTATCTC-CT--AAT-CGCCTCA-------------TCCA-----------A--TTCATTTTTG----------A--------C-----CCTTTGCCTACTTTTAGAACTTTGAAAAAGAC |
| droMoj3 | GAATCTAAACTAAATGTAGGTATAGT-AA--TAGATG----AGTTTTTCTTTTAT------------------------TTT---------A-A--------C-----CTTTGGCCCGCTTGCAGAACTTTGAGAAGGAC |
| droGri2 | GAATCCAAACTAAATGTGAGATACCTCTA-AAACAAA-T-CA-------------TAAA----GATTTTGA--TGTATTGAGA--------------------------TTATGTACATTTGCAGAACTTTGAGAAAGAC |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
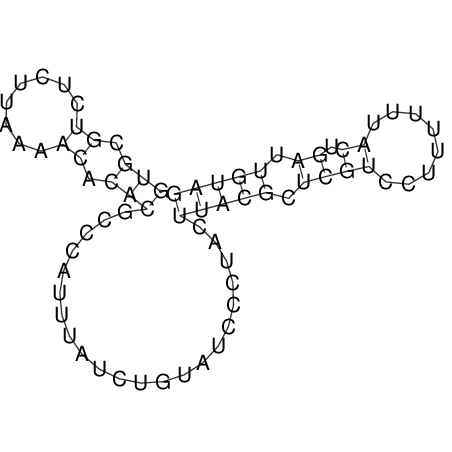
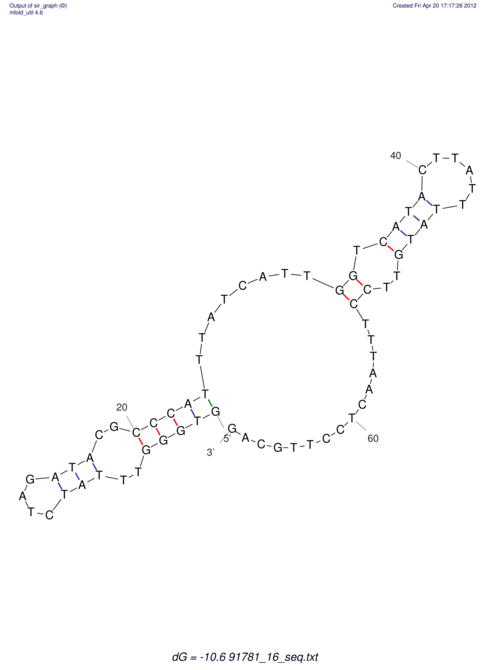
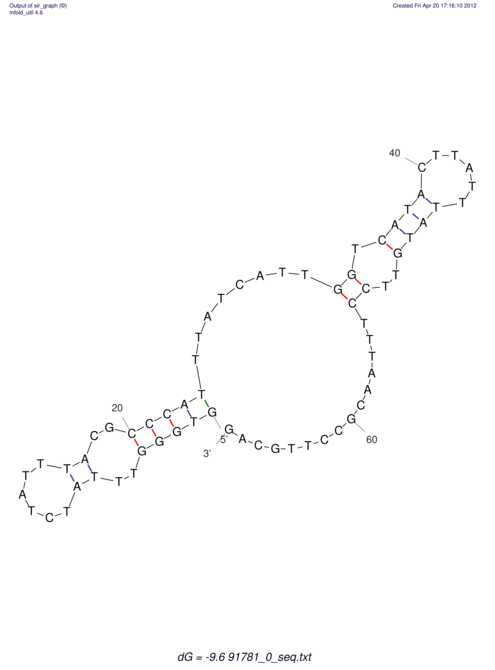
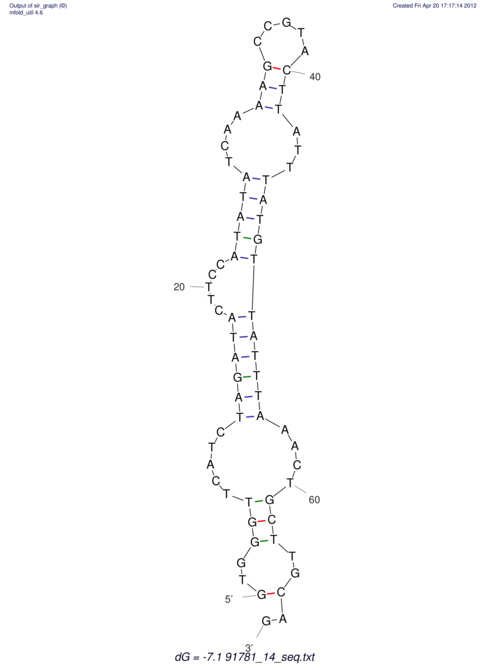
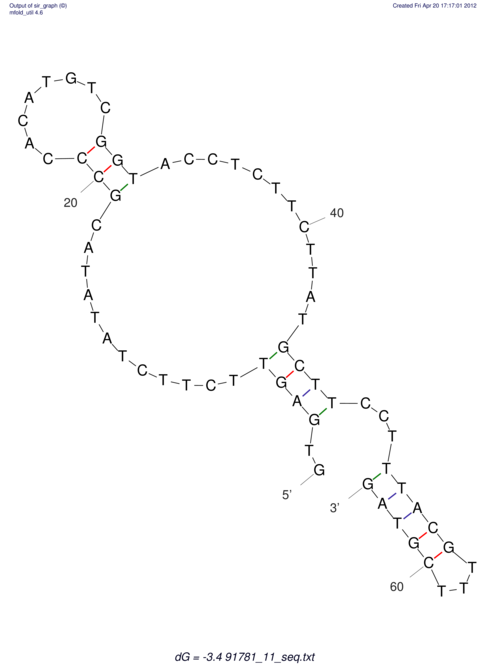
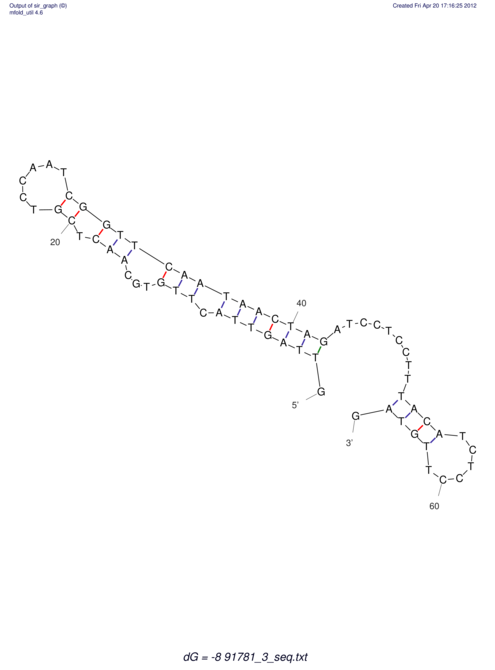
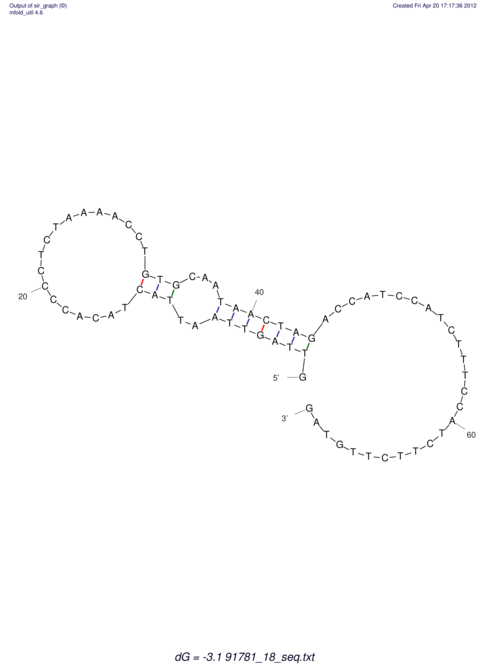
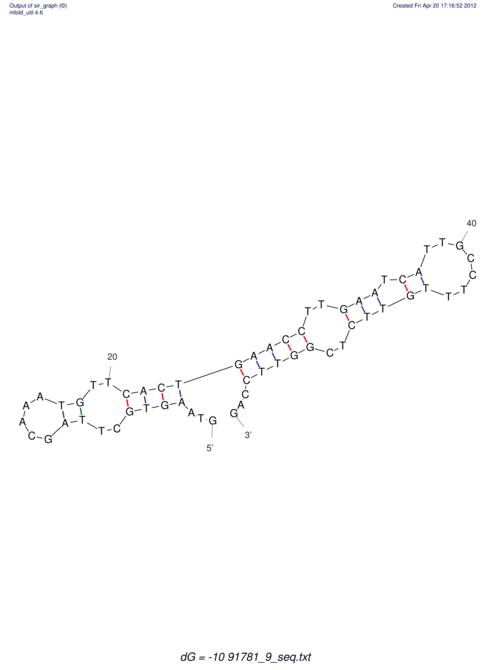
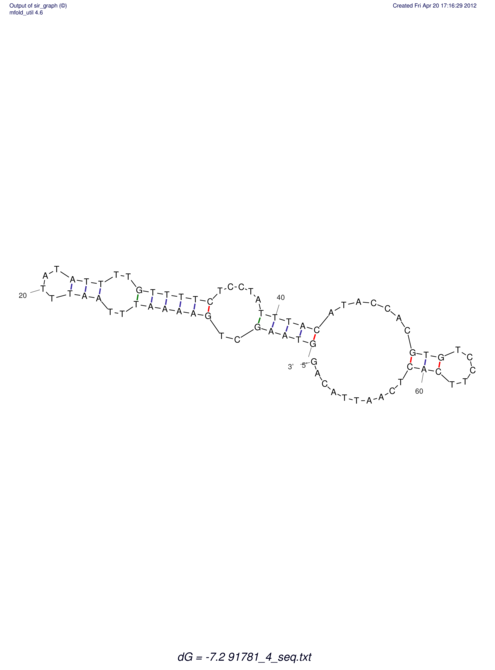
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
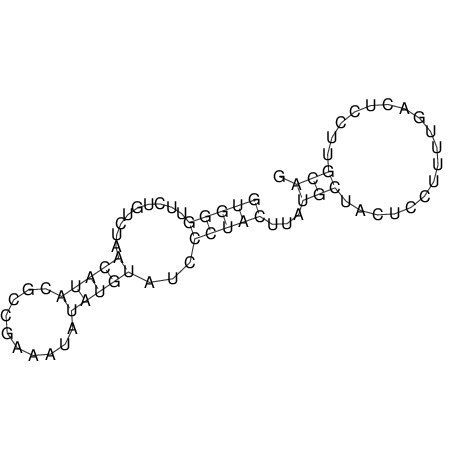
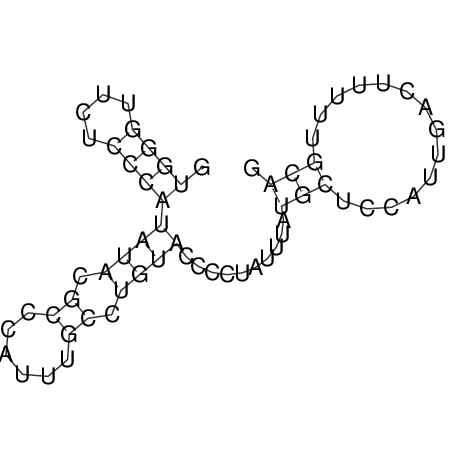
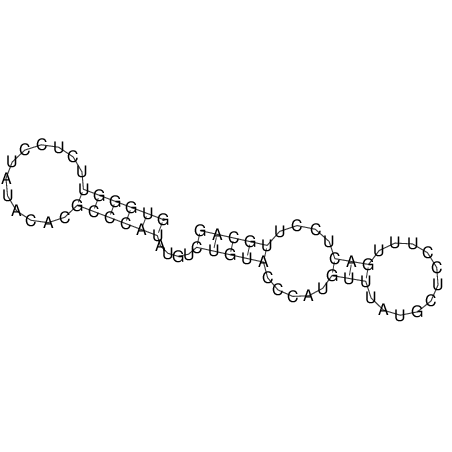
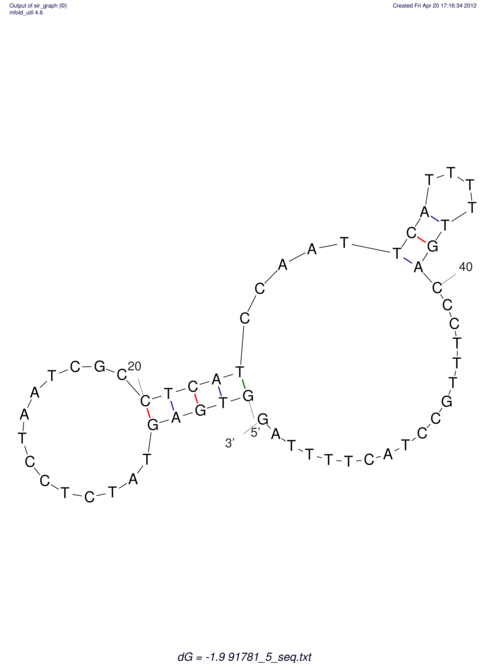
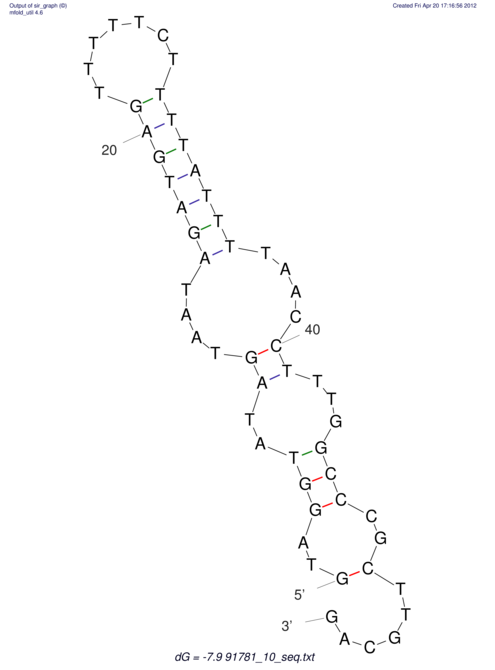
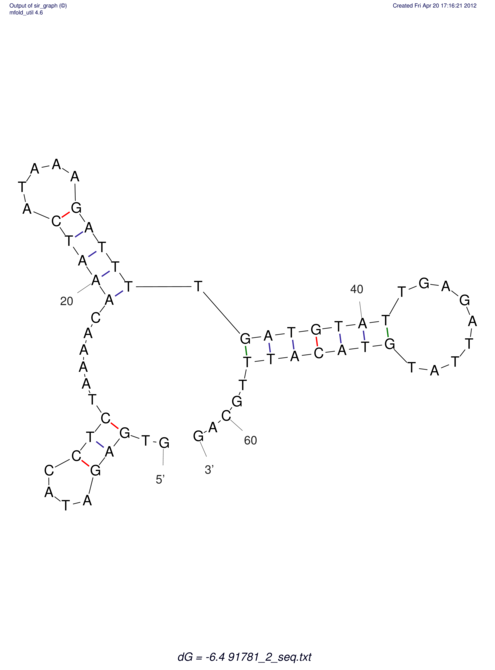
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:17:45 EDT