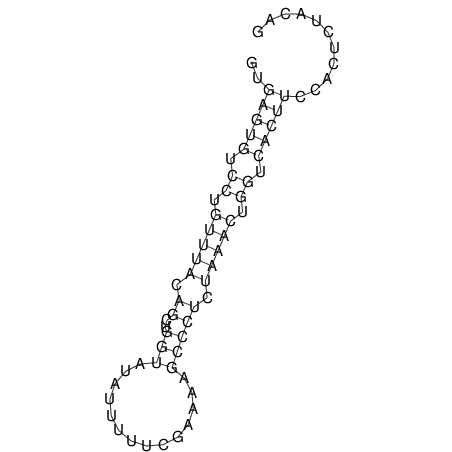
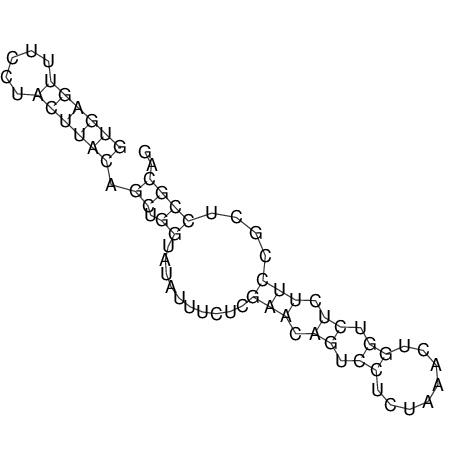
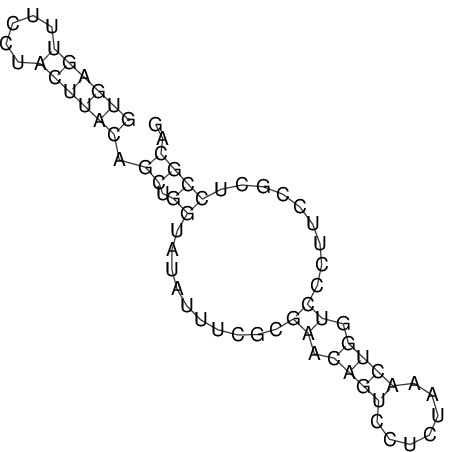
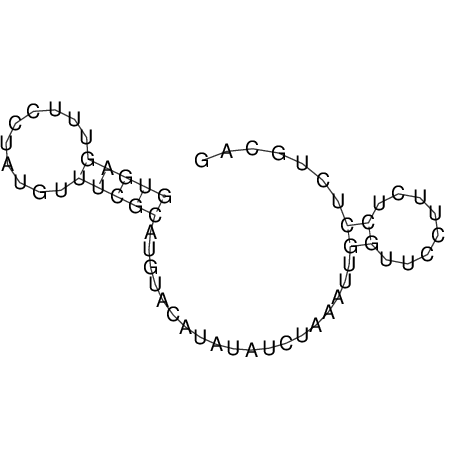



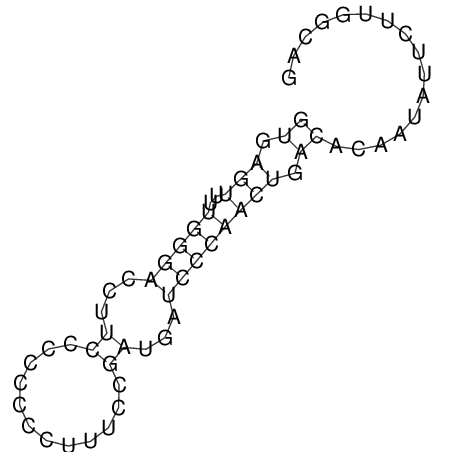
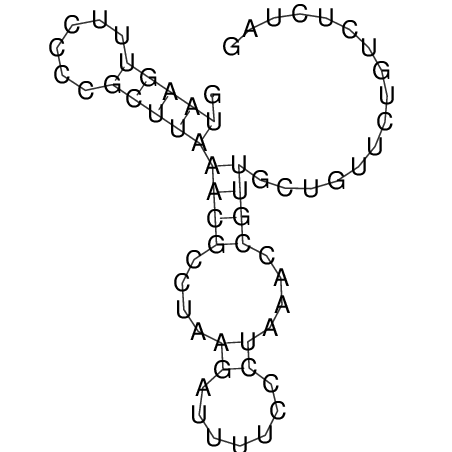
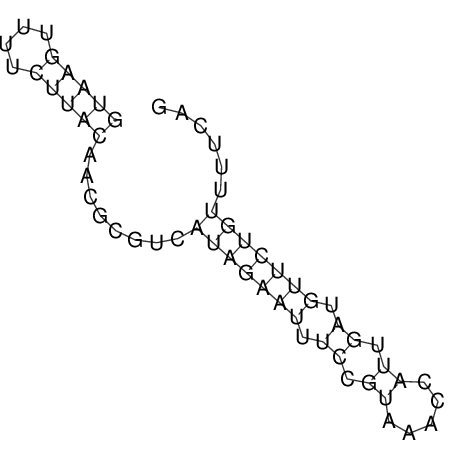

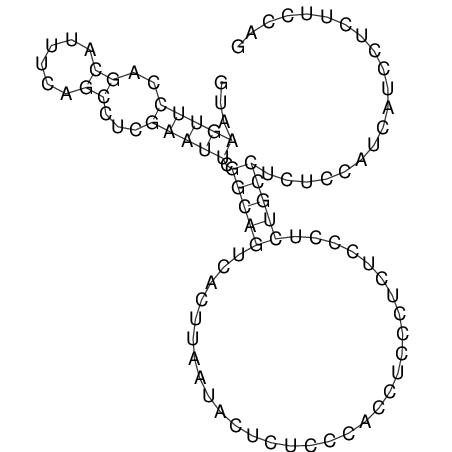


View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
dme-mir-3641 |
chrX:2065119-2065187 + |
Known_mirbase |
|---|---|---|
| [View on miRBase] | [View on UCSC Genome Browser {Cornell Mirror}] | tailed_mirtron |
Legend:
mature strand at position 2065119-2065138| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chrX:2065104-2065202 + | 1 | GGCGAACTTTTTAATGTGAGTGTCCT-------G--------------------------------------------------------------------TTTACAGCTGGTATATTTTTCGAAAAGCCCTCTA-----AACTGGT-CACTTCCACTCTACAGATCTATAACTACATA | |
| droSim1 | chrX:1441441-1441539 + | LASTZ | 5 | GGCGAACTGTTCAATGTGAGTTTCCT-------A--------------------------------------------------------------------CTTACAGCTGGTATATTTCTCGAACAGTCCTCTA-----AACTGGT-CTCTTCCGCTCCGCAGATCTACAACTACATA |
| droSec1 | super_10:1848730-1848828 + | LASTZ | 5 | GGCGAACTGTTCAATGTGAGTTTCCT-------A--------------------------------------------------------------------CTTACAGCTGGTATATTTCGCGAACAGTCCTCTA-----AACTGGT-CCCTTCCGCTCCGCAGATCTACAACTACATA |
| droYak2 | chrX:6259202-6259289 - | LASTZ | 5 | GGCGAACTCTTCAATGTGAGTTTCCT-------A--------------------------------------------------------------------TGTTTCGCATGTACATA-----------TATCTA-----AATTGGT-TCCTTCTCCTCTGCAGATCTACAACTACATA |
| droEre2 | scaffold_4644:2028284-2028382 + | LASTZ | 5 | GGCGAACTGTTCAATGTGAGTATCCT-------G--------------------------------------------------------------------TGATCCGCTTGTACTTATCTCGAGCAGTTCTCTA-----AACTGGT-CCCTGATCCTCTGCAGATCTACAACTACATA |
| droEug1 | scf7180000409093:49412-49503 - | LASTZ | 5 | GGGGAGCTGTTCAATGTGAGTTATTTGA------G---------------------------------------------------------------------TTATATTCCTG-------CTTATAACCCTCTA-----AATTGGA-CTTGTTGGTCTTGCAGATCTACAACTATATA |
| droBia1 | scf7180000301760:2041662-2041749 + | LASTZ | 5 | GGCGAGCTGTTCAACGTGAGTTCCCT-------T--------------------------------------------------------------------GCCCTCGCCGGCCTCTCCCTCAAGTAATCATCC-----------------ACCCTGCCCGCAGATCTACAACTACATA |
| droTak1 | scf7180000415191:145537-145623 + | LASTZ | 5 | GGCGAGCTGTTCAATGTGAGTTTCTTGG------G-----------------------------------------------------------------T-TTTCCAGGTGACCTAGTGACT------------G-----AGTGGG----CTACTATCTTACAGATCTACAACTACATA |
| droEle1 | scf7180000491001:1609052-1609151 + | LASTZ | 5 | GGCGAGCTGTTCAATGTGAGTAGCGC-----------------TCGGTTGCCCCCTAATCCTGCATTCCTTATCCTAATCCTTAT--------------------------------------------------------------C-CTCCCACCCCCCGCAGATCTACAACTACATC |
| droRho1 | scf7180000779506:128106-128197 + | LASTZ | 5 | GGCGAGCTGTTCAATGTGAGTGGTTT-------------------------------------------------------------------------------ACAGTC----CTTGTCCCTAGTGGCCCTCTA-ATCC--CTTGT--GTGTTCCTCCTGCAGATCTACAACTACATC |
| droFic1 | scf7180000454072:3187079-3187168 + | LASTZ | 5 | GGCGAGCTGTTCAACGTGAGTTCCTCCG------G-------------------------------------------------------CATCCCTTCTTC--------------------TTCCTGCTCCTCTA-----ATCCGGC-CA---CTTGCTTGCAGATCTACAACTACATC |
| droKik1 | scf7180000302696:632884-632975 + | LASTZ | 5 | GGCGAGCTGTTCAATGTGAGTTTTGGGA------C---------------------------------------------------------------------CTTCCCCCCCC-------TTTCCGATGATCCC-----AACTGAC-ACAATATTCTTGGCAGATCTACAACTACATC |
| droAna3 | scaffold_13117:1070684-1070773 - | LASTZ | 5 | GGCGAGCTCTTCAACGTAAGTTTCCCCG------C--------------------------------------------------------------------TTAAACGCCTAA----------GATTTTCCCTA-----AACCGTT-GCTGTTCTGTCTCTAGATCTACAACTATATC |
| droBip1 | scf7180000396425:1044645-1044733 + | LASTZ | 5 | GGTGAGCTCTTCAATGTAAGTTTTCT-------T--------------------------------------------------------------------ACAACGCGTCATA----------GAATTTCCGTA-----AACCATT-GATGTTCTGTTTTCAGATCTACAACTATATC |
| dp4 | chrXL_group1e:3790116-3790235 - | LASTZ | 5 | GGCGAGTTGTTCAACGTAAGTTCCAGCATTTCA-GCCTCGAATTCGGCAGTCACTTGA------------------------TACTCTCCCACCTCCCTCTC--------------------CCTC-------------TG--CCTCTCCATCATCCTCTTCCAGATCTACAACTACATC |
| droPer1 | super_17:723995-724114 - | LASTZ | 5 | GGCGAGTTGTTCAACGTAAGTTCCAGCATTTCA-GCCTCGAATTCGGCAGTCACTTAA------------------------TACTCTCCCACCTCCCTCTC--------------------CCTC-------------TG--CCTCTCCATCATCCTCTTCCAGATCTACAACTACATC |
| droWil1 | scaffold_181096:7421152-7421244 - | LASTZ | 5 | GGCGAGCTTTTCAACGTGAGTTACTCAG------T------------------------CCAAAGAAACTGGTTTTAGTTATGAT-----------------------------------------TAA--------------CTCA--CTTTTTTTTGTTATAGATTTACAACTATATA |
| droVir3 | scaffold_12928:7113520-7113613 + | LASTZ | 5 | GGCGAGTTGTTCAACGTAAGTCGATG-------A--------------------------------------------------------------------CCTCGCGTATGTGGG-----CGTACTTTCACTGA-----CGCCTGC-CTCTTTGTCTATTCAGATTTACAACTACATT |
| droMoj3 | scaffold_6328:2787435-2787524 + | LASTZ | 5 | GGGGAGTTGTTCAACGTAAATTCTAT-----------------------------------------------------------------------------TCTCACTTAACACAAGAATC----AAA--TCTC-----ATCTAAT--TTTTCGCTCGTTTAGATTTACAACTACATT |
| droGri2 | scaffold_14853:3491672-3491772 - | LASTZ | 5 | GGAGAGTTGTTTAATGTAAGCTAAA---------C-------TTCGCCCATCCT-------------------------------------------------CTCCAGCTGTTTGG-----TTAA-----CTTTCAACTG--TTTGT--CCTCCATCTCCTCAGATTTACAACTACATT |
| Species | Alignment |
| dm3 | GGCGAACTTTTTAATGTGAGTGTCCT-------G--------------------------------------------------------------------TTTACAGCTGGTATATTTTTCGAAAAGCCCTCTA-----AACTGGT-CACTTCCACTCTACAGATCTATAACTACATA |
| droSim1 | GGCGAACTGTTCAATGTGAGTTTCCT-------A--------------------------------------------------------------------CTTACAGCTGGTATATTTCTCGAACAGTCCTCTA-----AACTGGT-CTCTTCCGCTCCGCAGATCTACAACTACATA |
| droSec1 | GGCGAACTGTTCAATGTGAGTTTCCT-------A--------------------------------------------------------------------CTTACAGCTGGTATATTTCGCGAACAGTCCTCTA-----AACTGGT-CCCTTCCGCTCCGCAGATCTACAACTACATA |
| droYak2 | GGCGAACTCTTCAATGTGAGTTTCCT-------A--------------------------------------------------------------------TGTTTCGCATGTACATA-----------TATCTA-----AATTGGT-TCCTTCTCCTCTGCAGATCTACAACTACATA |
| droEre2 | GGCGAACTGTTCAATGTGAGTATCCT-------G--------------------------------------------------------------------TGATCCGCTTGTACTTATCTCGAGCAGTTCTCTA-----AACTGGT-CCCTGATCCTCTGCAGATCTACAACTACATA |
| droEug1 | GGGGAGCTGTTCAATGTGAGTTATTTGA------G---------------------------------------------------------------------TTATATTCCTG-------CTTATAACCCTCTA-----AATTGGA-CTTGTTGGTCTTGCAGATCTACAACTATATA |
| droBia1 | GGCGAGCTGTTCAACGTGAGTTCCCT-------T--------------------------------------------------------------------GCCCTCGCCGGCCTCTCCCTCAAGTAATCATCC-----------------ACCCTGCCCGCAGATCTACAACTACATA |
| droTak1 | GGCGAGCTGTTCAATGTGAGTTTCTTGG------G-----------------------------------------------------------------T-TTTCCAGGTGACCTAGTGACT------------G-----AGTGGG----CTACTATCTTACAGATCTACAACTACATA |
| droEle1 | GGCGAGCTGTTCAATGTGAGTAGCGC-----------------TCGGTTGCCCCCTAATCCTGCATTCCTTATCCTAATCCTTAT--------------------------------------------------------------C-CTCCCACCCCCCGCAGATCTACAACTACATC |
| droRho1 | GGCGAGCTGTTCAATGTGAGTGGTTT-------------------------------------------------------------------------------ACAGTC----CTTGTCCCTAGTGGCCCTCTA-ATCC--CTTGT--GTGTTCCTCCTGCAGATCTACAACTACATC |
| droFic1 | GGCGAGCTGTTCAACGTGAGTTCCTCCG------G-------------------------------------------------------CATCCCTTCTTC--------------------TTCCTGCTCCTCTA-----ATCCGGC-CA---CTTGCTTGCAGATCTACAACTACATC |
| droKik1 | GGCGAGCTGTTCAATGTGAGTTTTGGGA------C---------------------------------------------------------------------CTTCCCCCCCC-------TTTCCGATGATCCC-----AACTGAC-ACAATATTCTTGGCAGATCTACAACTACATC |
| droAna3 | GGCGAGCTCTTCAACGTAAGTTTCCCCG------C--------------------------------------------------------------------TTAAACGCCTAA----------GATTTTCCCTA-----AACCGTT-GCTGTTCTGTCTCTAGATCTACAACTATATC |
| droBip1 | GGTGAGCTCTTCAATGTAAGTTTTCT-------T--------------------------------------------------------------------ACAACGCGTCATA----------GAATTTCCGTA-----AACCATT-GATGTTCTGTTTTCAGATCTACAACTATATC |
| dp4 | GGCGAGTTGTTCAACGTAAGTTCCAGCATTTCA-GCCTCGAATTCGGCAGTCACTTGA------------------------TACTCTCCCACCTCCCTCTC--------------------CCTC-------------TG--CCTCTCCATCATCCTCTTCCAGATCTACAACTACATC |
| droPer1 | GGCGAGTTGTTCAACGTAAGTTCCAGCATTTCA-GCCTCGAATTCGGCAGTCACTTAA------------------------TACTCTCCCACCTCCCTCTC--------------------CCTC-------------TG--CCTCTCCATCATCCTCTTCCAGATCTACAACTACATC |
| droWil1 | GGCGAGCTTTTCAACGTGAGTTACTCAG------T------------------------CCAAAGAAACTGGTTTTAGTTATGAT-----------------------------------------TAA--------------CTCA--CTTTTTTTTGTTATAGATTTACAACTATATA |
| droVir3 | GGCGAGTTGTTCAACGTAAGTCGATG-------A--------------------------------------------------------------------CCTCGCGTATGTGGG-----CGTACTTTCACTGA-----CGCCTGC-CTCTTTGTCTATTCAGATTTACAACTACATT |
| droMoj3 | GGGGAGTTGTTCAACGTAAATTCTAT-----------------------------------------------------------------------------TCTCACTTAACACAAGAATC----AAA--TCTC-----ATCTAAT--TTTTCGCTCGTTTAGATTTACAACTACATT |
| droGri2 | GGAGAGTTGTTTAATGTAAGCTAAA---------C-------TTCGCCCATCCT-------------------------------------------------CTCCAGCTGTTTGG-----TTAA-----CTTTCAACTG--TTTGT--CCTCCATCTCCTCAGATTTACAACTACATT |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
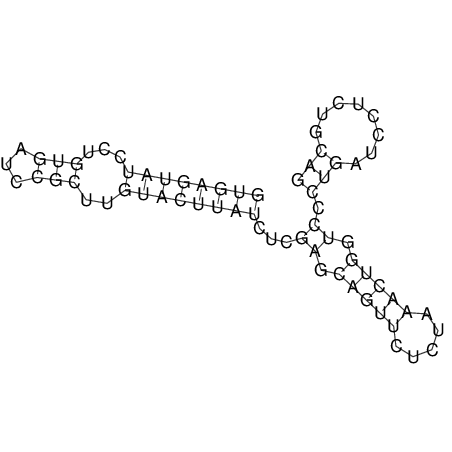
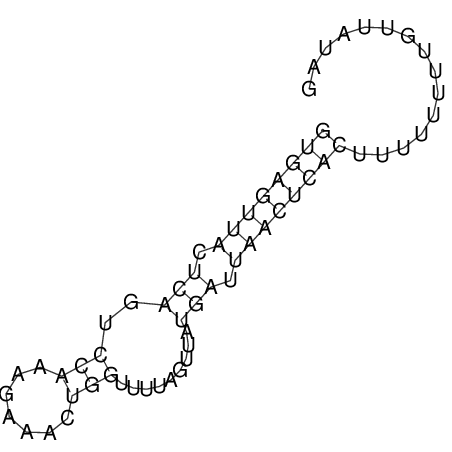
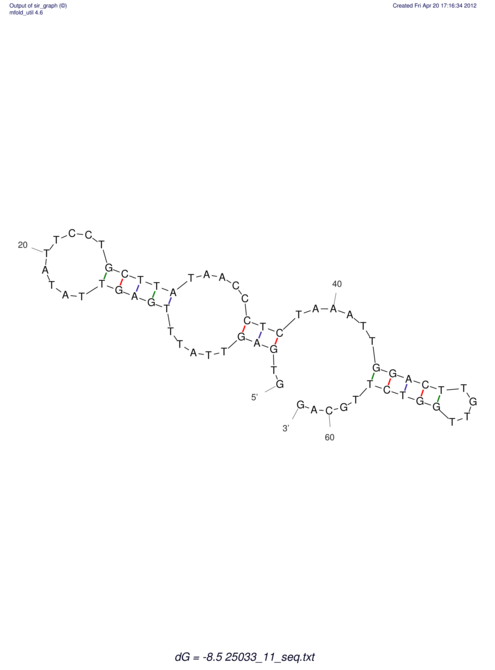
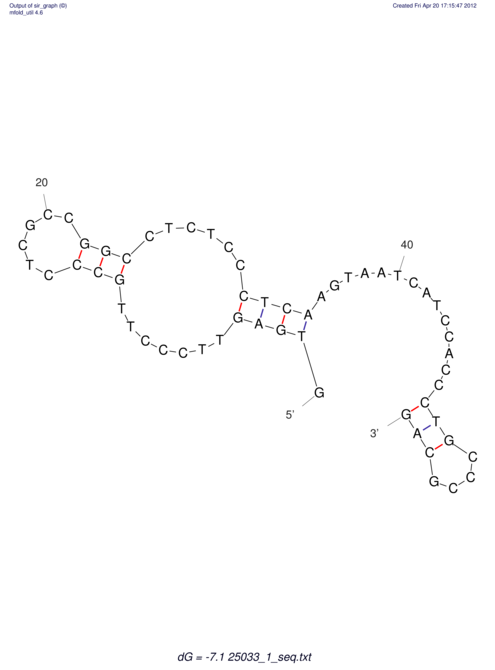
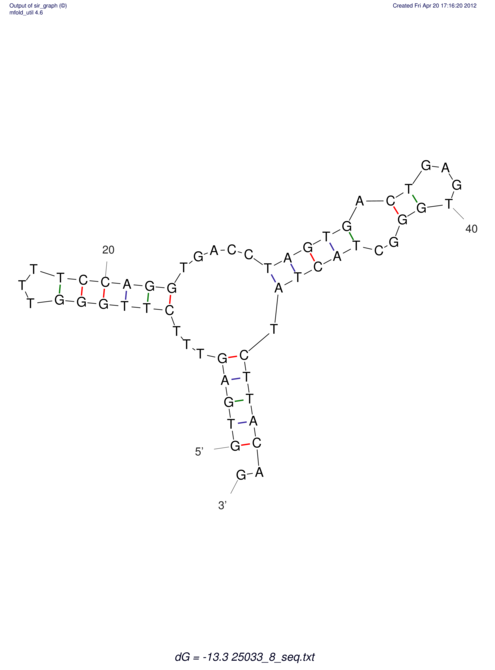
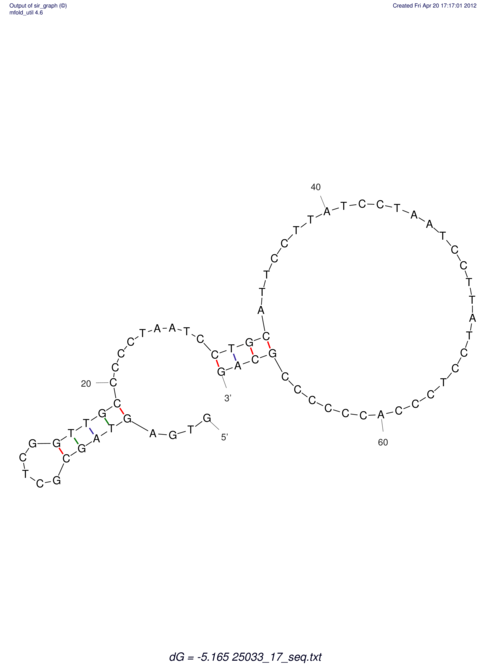
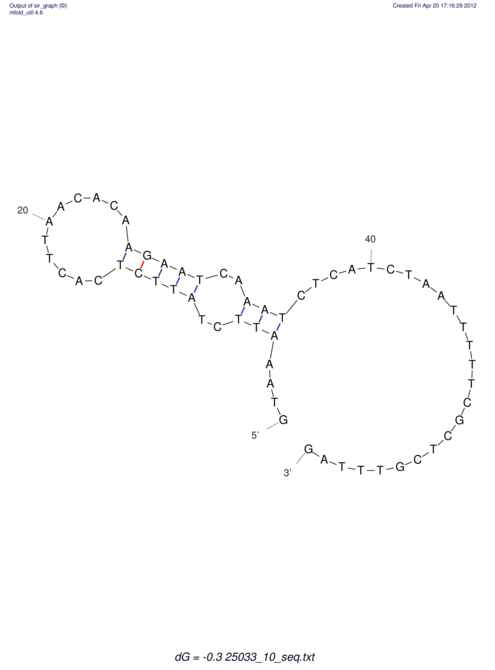
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
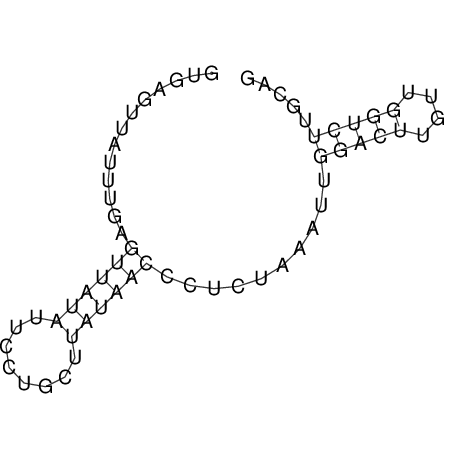
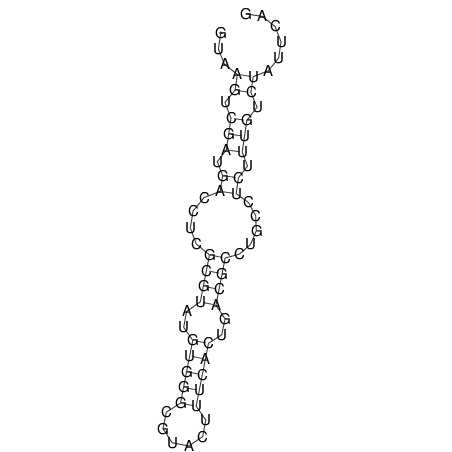
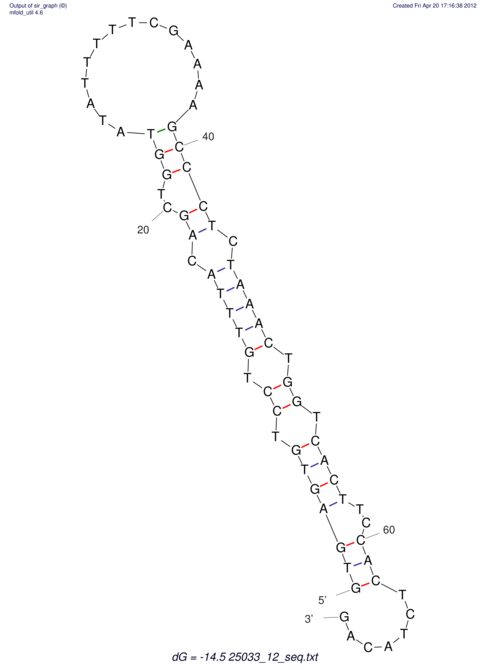
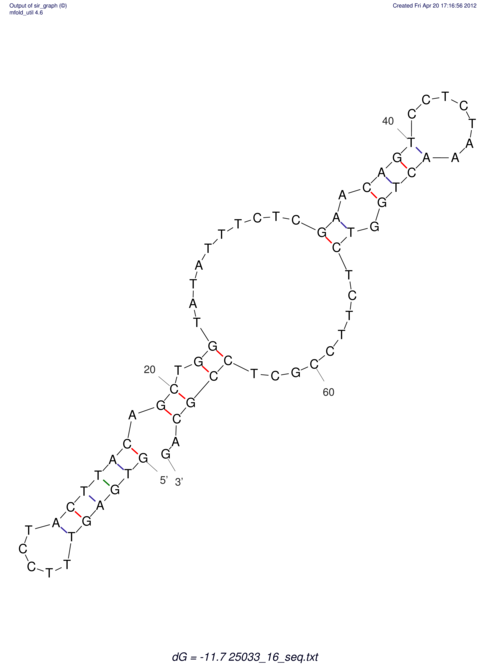
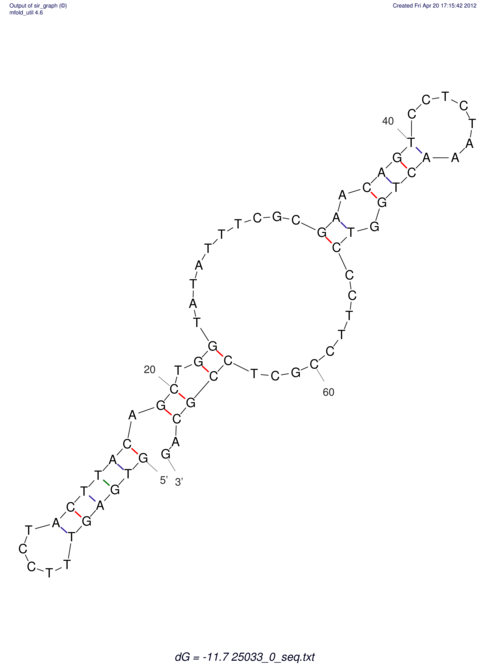
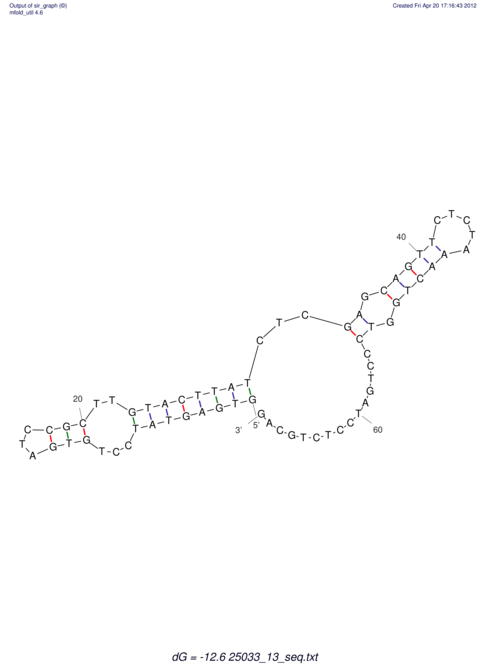
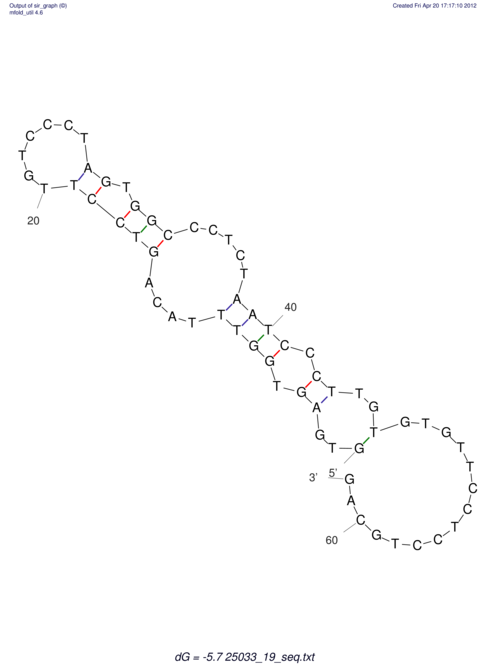
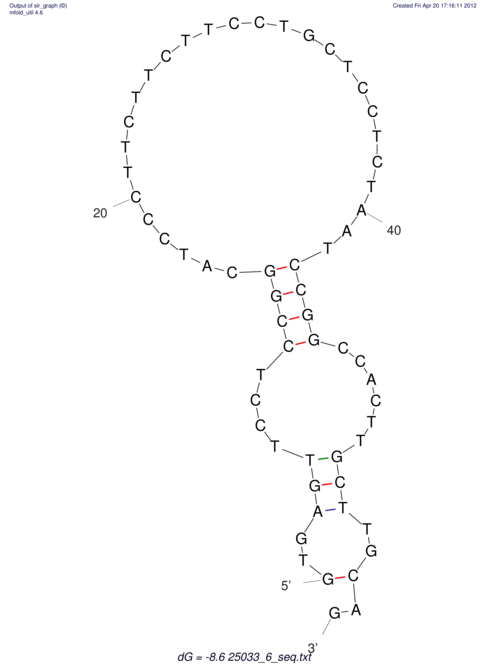
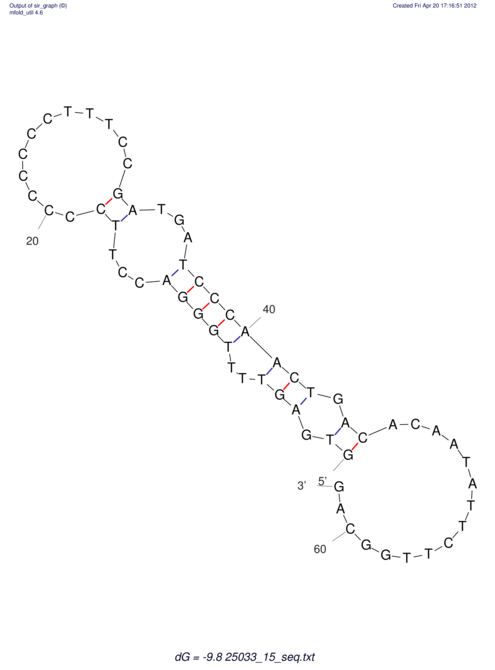
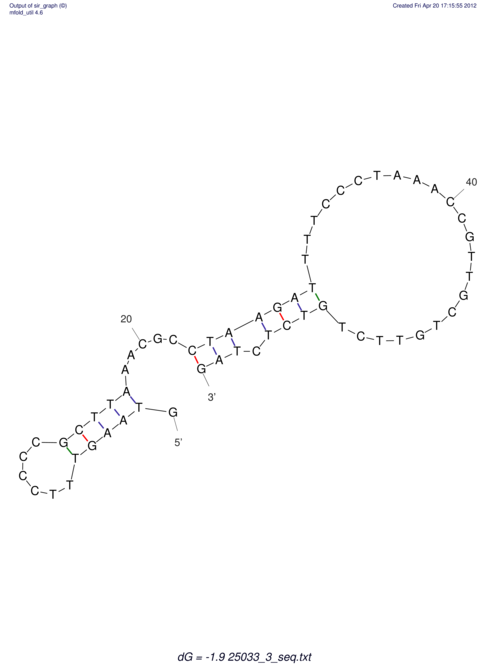
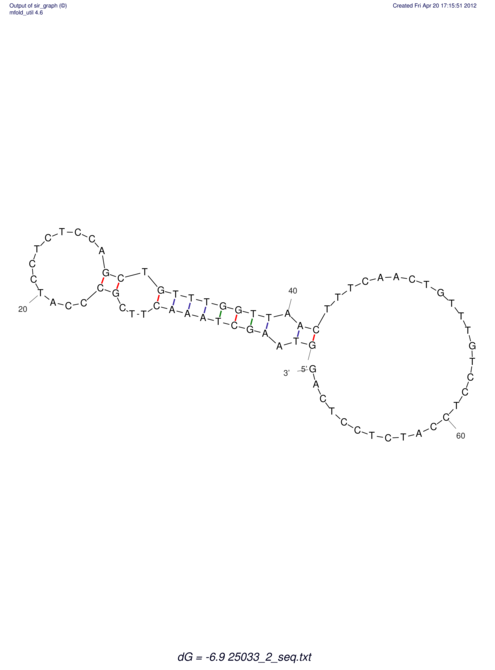
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
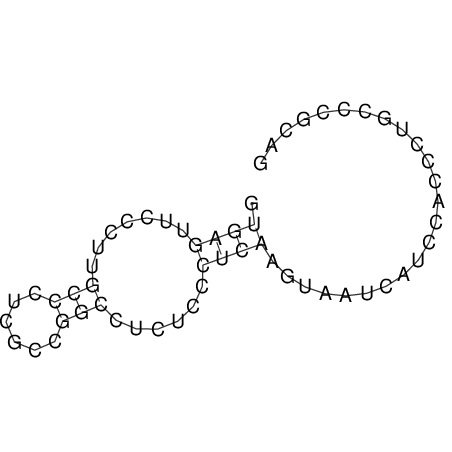
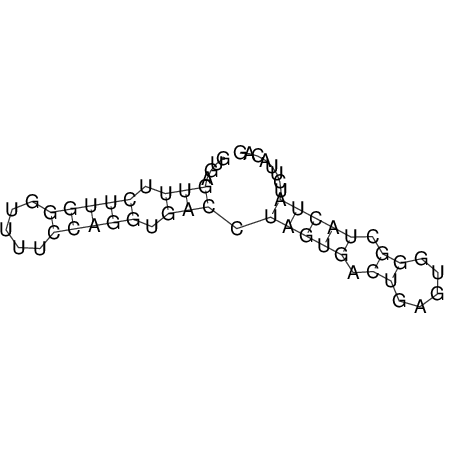
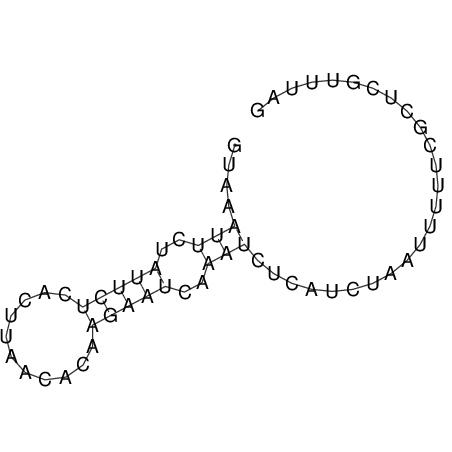
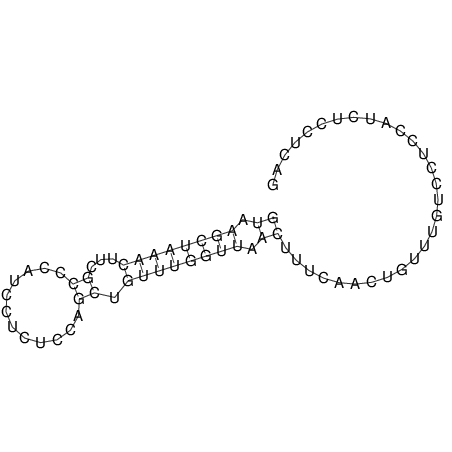
| Species | Alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:17:15 EDT