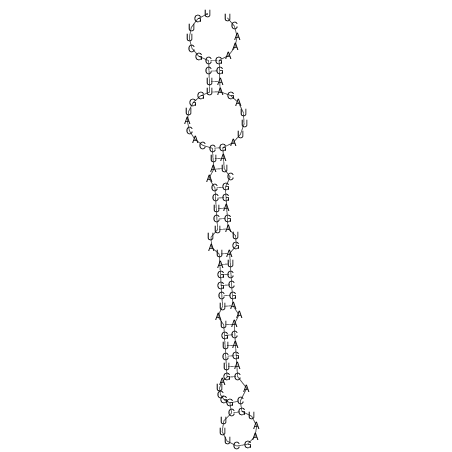
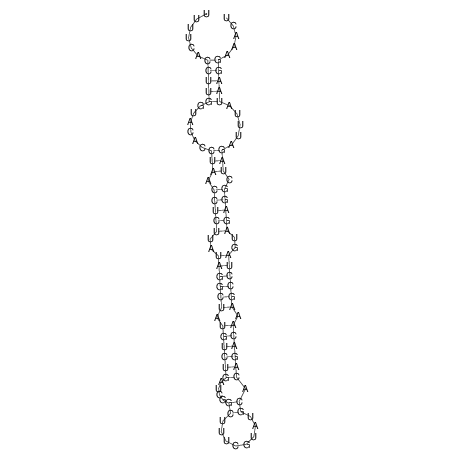
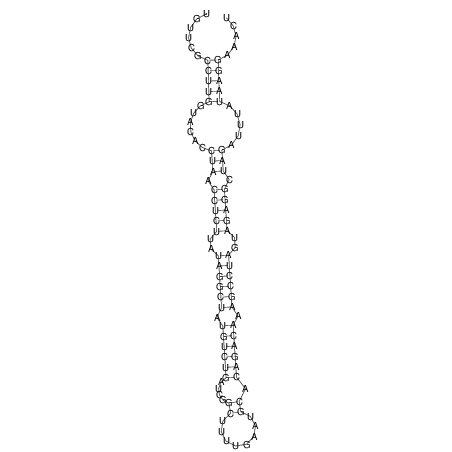
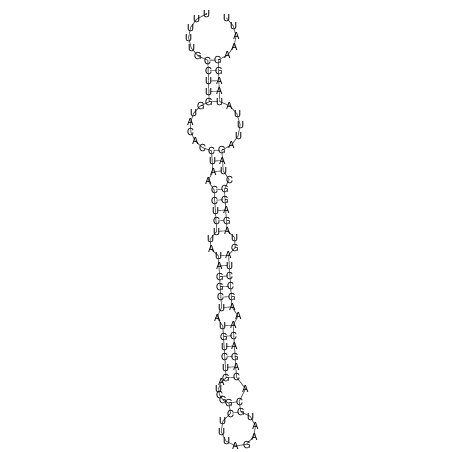
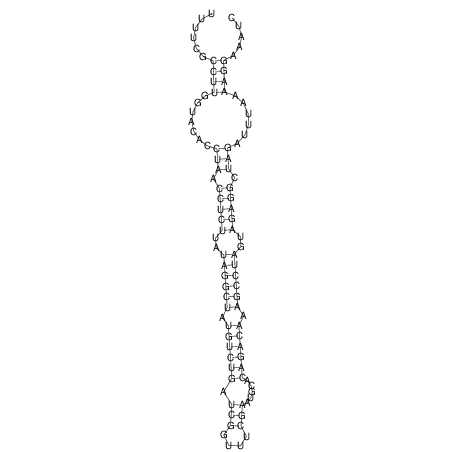
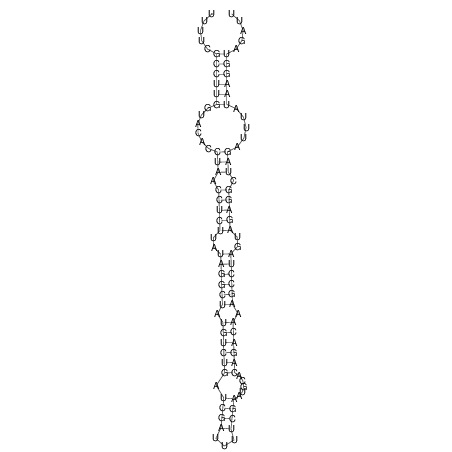
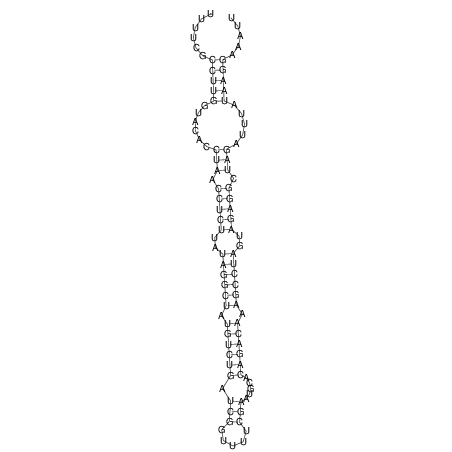


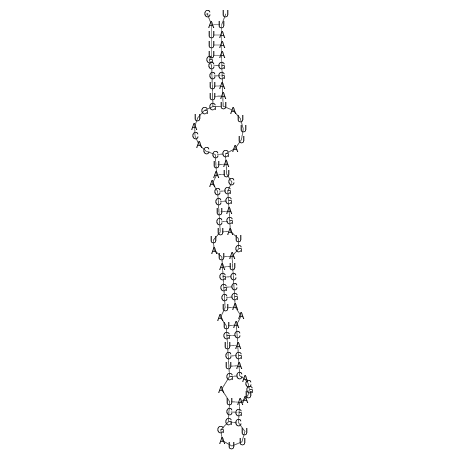
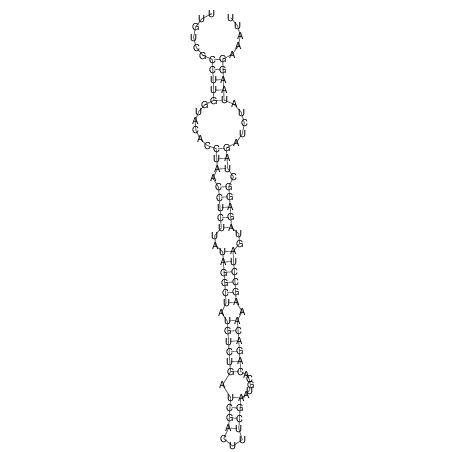

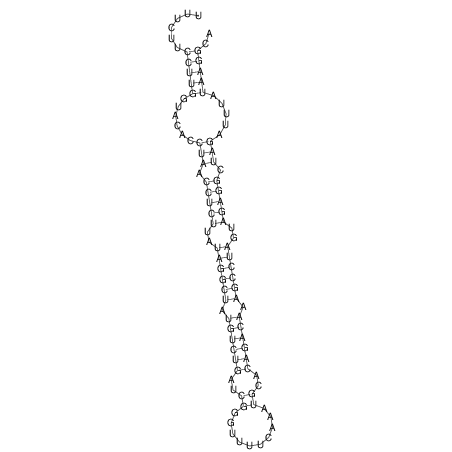
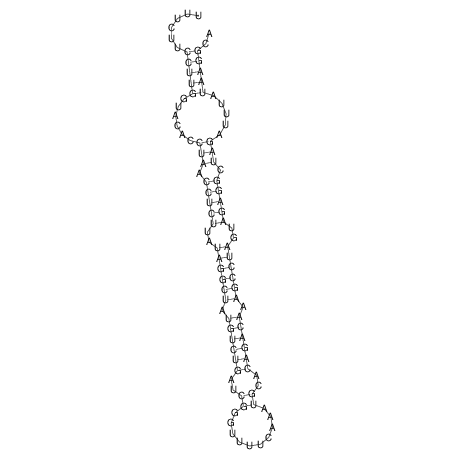
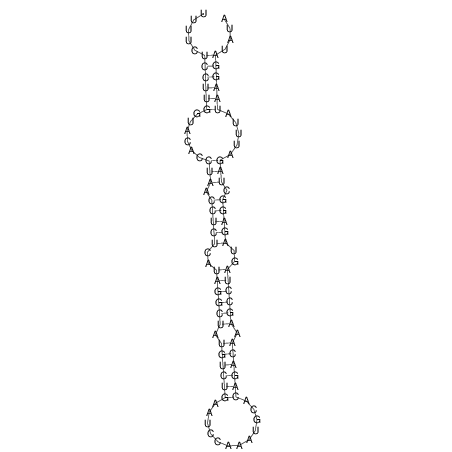
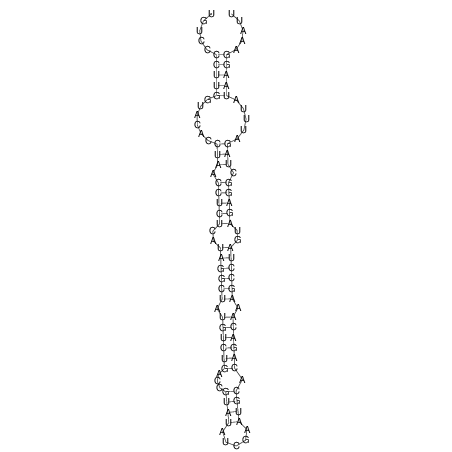
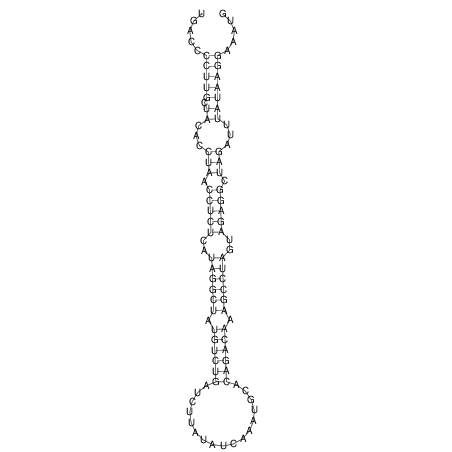



View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
dme-mir-282-as |
chr3L:3251022-3251118 - |
Candidate |
|---|---|---|
| [View on UCSC Genome Browser {Cornell Mirror}] | intergenic |
Legend:
mature strand at position 3251036-3251057| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr3L:3251007-3251133 - | 1 | TCCGTTTT------------------CGCTGATTGTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAGAAGGAAA-CTATA--------G-TTAAAGAAC-----------GG------------- | |
| droSim1 | chr3L:2784381-2784507 - | LASTZ | 4 | TCCGTTTT------------------CGCTGATTGTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAAAAGGAAA-CTTTA--------G-TTTAAGAAC-----------GG------------- |
| droSec1 | super_2:3244745-3244871 - | LASTZ | 3 | TCCGTTTT------------------CGCTGATTGTTCACCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAAAAGGAAA-CTTTA--------T-TTTAAGAAC-----------GG------------- |
| droYak2 | chr3L:3795918-3796050 - | LASTZ | 3 | TCCGTTTT------------------CGCTGATTTTTCACCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGTA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-CTGAT--A-TAACGGATATAGAAC-----------GA------------- |
| droEre2 | scaffold_4784:5933708-5933834 - | LASTZ | 4 | TCCGTTTT------------------CGCTGCTTGTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTTGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-CTGTG--------G-TGATAGAAC-----------GA------------- |
| droEug1 | scf7180000409466:975797-975923 - | LASTZ | 4 | TCCGTTTT------------------TGCTGACTTTTTGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTAGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTCTC--------G-GTGTATAAC-----------GG------------- |
| droBia1 | scf7180000302428:2604017-2604144 - | LASTZ | 4 | TCCGATTC------------------CGATGATTTTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG--TTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAAAAGGAAA-TCTGC----------ACTTAGAAG-----------GGTAC---------- |
| droTak1 | scf7180000415378:123710-123845 + | LASTZ | 4 | TCCGTTTTC-----------------TGATGATTTTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGA-TTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGTAGATTG-CAGATTCTCG-ATTTAGAAT-----------GG------------- |
| droEle1 | scf7180000491249:2016913-2017039 + | LASTZ | 4 | GCCGTTTT------------------TGATGCTTTTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-TTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTCTT--------G-AGTTAGAAC-----------AG------------- |
| droRho1 | scf7180000757706:21549-21675 + | LASTZ | 4 | TCCGTTTT------------------TGCTGATTTTTCGCCTTTGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-TTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTATT--------G-GATTAGAAC-----------GG------------- |
| droFic1 | scf7180000454105:1571232-1571357 - | LASTZ | 4 | GCCCTCTT------------------CGCTGCTCTTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAAAAGGAGA-TTCTC--------G-G-CCAGAAC-----------GG------------- |
| droKik1 | scf7180000302391:353031-353159 - | LASTZ | 4 | TCCGCCTTT-----------------CGCTGCACATTTGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-ATTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTCTC--------GGATTCAGAAC-----------GG------------- |
| droAna3 | scaffold_13337:7556582-7556720 - | LASTZ | 3 | TCCGTTTTTG----------------TGATGTCTTGTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGA-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATCTATAAGGAAA-TTTTC-GATTTTCG-AATTAGAAC-----------GGCCA---------- |
| droBip1 | scf7180000395543:216902-217046 - | LASTZ | 4 | TCCGTTTTAA----------------TGATGACT-TTTGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATC--------GAA--AACACAGACAAAGCCTAGTAGAGGCTAGATCTATAAGGAAA-TTTTC--------G-ATTTAGAACGTAGAGCATATTGCCACGCCCACACG |
| dp4 | chrXR_group8:3299974-3300083 + | LASTZ | 3 | TCCGCTTT------------------TGATGCATTTCTTCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGGGTTTTCAAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGCA---------------------------------------------------- |
| droPer1 | super_65:103931-104040 + | LASTZ | 3 | TCCGCTTT------------------TGATGCATTTCTTCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGGGTTTTCAAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGCA---------------------------------------------------- |
| droWil1 | scaffold_181136:1999668-1999787 - | LASTZ | 5 | T--------------------------GTTGTATTTTCTCCTTGGTACACCTAACCTCTCATAGGCTATGTCTGAA------TCCAAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGATA-TATTA-ATATT------ACATAAA-----GGAA--AA------------- |
| droVir3 | scaffold_12758:910696-910814 + | LASTZ | 4 | --CGCCCT------------------CGCTGCTT-GTCCCCTTGGTACACCTAACCTCTCATAGGCTATGTCTGACCGT-ATATCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTT--------------GTCGGAC-----------GC------------- |
| droMoj3 | scaffold_6680:1963284-1963406 + | LASTZ | 3 | TCT----C------------------TGCTGTT-TGACCCCTTGCTACACCTAACCTCTCATAGGCTATGTCTGATCTT-ATATCAAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TGCAC--------CGCAAAAAAAA-----------AT------------- |
| droGri2 | scaffold_15110:607074-607202 - | LASTZ | 4 | TCATTTTTAGTCCCCGACAGCAGCCTTGTTGCTTTGATCCATTTGTACACCTAACCTCTCATAGGCTATGTCTGATTGA-ATTGAAAAAGTGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAA---------------------------------------------------- |
| Species | Alignment |
| dm3 | TCCGTTTT------------------CGCTGATTGTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAGAAGGAAA-CTATA--------G-TTAAAGAAC-----------GG------------- |
| droSim1 | TCCGTTTT------------------CGCTGATTGTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAAAAGGAAA-CTTTA--------G-TTTAAGAAC-----------GG------------- |
| droSec1 | TCCGTTTT------------------CGCTGATTGTTCACCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAAAAGGAAA-CTTTA--------T-TTTAAGAAC-----------GG------------- |
| droYak2 | TCCGTTTT------------------CGCTGATTTTTCACCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGTA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-CTGAT--A-TAACGGATATAGAAC-----------GA------------- |
| droEre2 | TCCGTTTT------------------CGCTGCTTGTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTTGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-CTGTG--------G-TGATAGAAC-----------GA------------- |
| droEug1 | TCCGTTTT------------------TGCTGACTTTTTGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTAGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTCTC--------G-GTGTATAAC-----------GG------------- |
| droBia1 | TCCGATTC------------------CGATGATTTTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG--TTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAAAAGGAAA-TCTGC----------ACTTAGAAG-----------GGTAC---------- |
| droTak1 | TCCGTTTTC-----------------TGATGATTTTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGA-TTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGTAGATTG-CAGATTCTCG-ATTTAGAAT-----------GG------------- |
| droEle1 | GCCGTTTT------------------TGATGCTTTTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-TTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTCTT--------G-AGTTAGAAC-----------AG------------- |
| droRho1 | TCCGTTTT------------------TGCTGATTTTTCGCCTTTGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-TTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTATT--------G-GATTAGAAC-----------GG------------- |
| droFic1 | GCCCTCTT------------------CGCTGCTCTTTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTAAAAGGAGA-TTCTC--------G-G-CCAGAAC-----------GG------------- |
| droKik1 | TCCGCCTTT-----------------CGCTGCACATTTGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGG-ATTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTCTC--------GGATTCAGAAC-----------GG------------- |
| droAna3 | TCCGTTTTTG----------------TGATGTCTTGTCGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGA-CTTTCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATCTATAAGGAAA-TTTTC-GATTTTCG-AATTAGAAC-----------GGCCA---------- |
| droBip1 | TCCGTTTTAA----------------TGATGACT-TTTGCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATC--------GAA--AACACAGACAAAGCCTAGTAGAGGCTAGATCTATAAGGAAA-TTTTC--------G-ATTTAGAACGTAGAGCATATTGCCACGCCCACACG |
| dp4 | TCCGCTTT------------------TGATGCATTTCTTCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGGGTTTTCAAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGCA---------------------------------------------------- |
| droPer1 | TCCGCTTT------------------TGATGCATTTCTTCCTTGGTACACCTAACCTCTTATAGGCTATGTCTGATCGGGTTTTCAAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGCA---------------------------------------------------- |
| droWil1 | T--------------------------GTTGTATTTTCTCCTTGGTACACCTAACCTCTCATAGGCTATGTCTGAA------TCCAAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGATA-TATTA-ATATT------ACATAAA-----GGAA--AA------------- |
| droVir3 | --CGCCCT------------------CGCTGCTT-GTCCCCTTGGTACACCTAACCTCTCATAGGCTATGTCTGACCGT-ATATCGAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TTT--------------GTCGGAC-----------GC------------- |
| droMoj3 | TCT----C------------------TGCTGTT-TGACCCCTTGCTACACCTAACCTCTCATAGGCTATGTCTGATCTT-ATATCAAA--TGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAAA-TGCAC--------CGCAAAAAAAA-----------AT------------- |
| droGri2 | TCATTTTTAGTCCCCGACAGCAGCCTTGTTGCTTTGATCCATTTGTACACCTAACCTCTCATAGGCTATGTCTGATTGA-ATTGAAAAAGTGCACAGACAAAGCCTAGTAGAGGCTAGATTTATAAGGAA---------------------------------------------------- |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
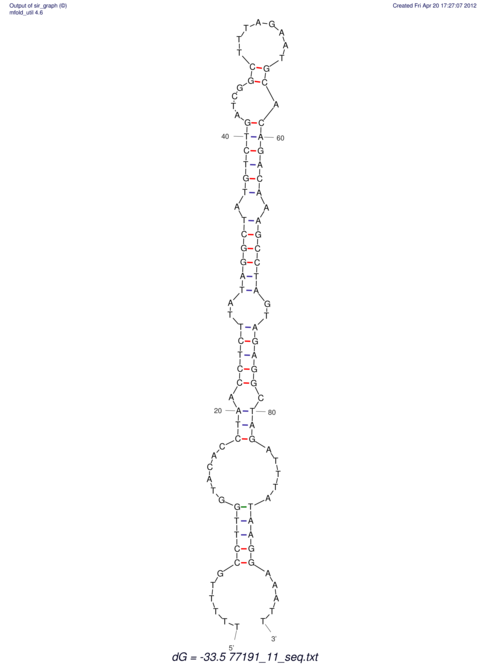
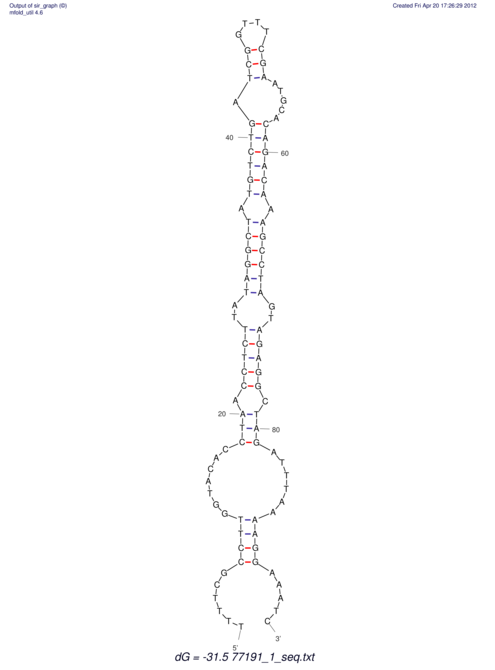
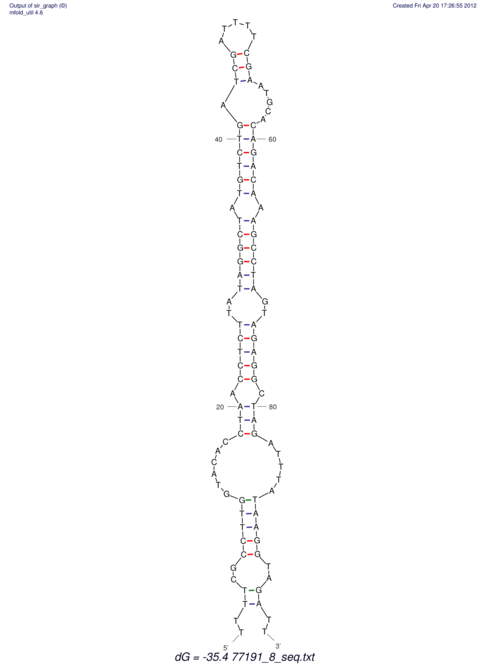
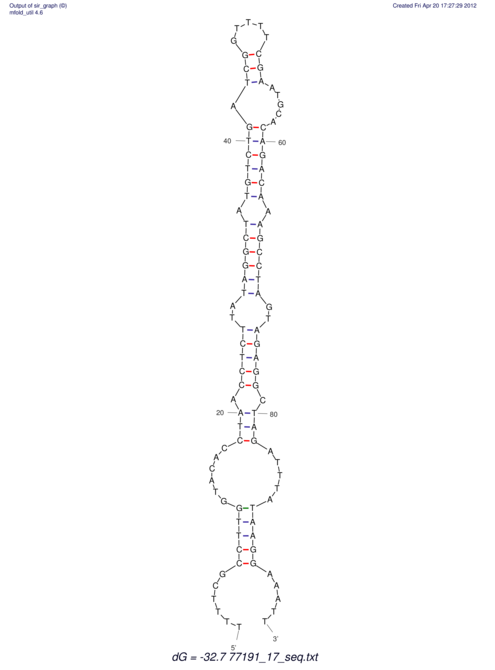
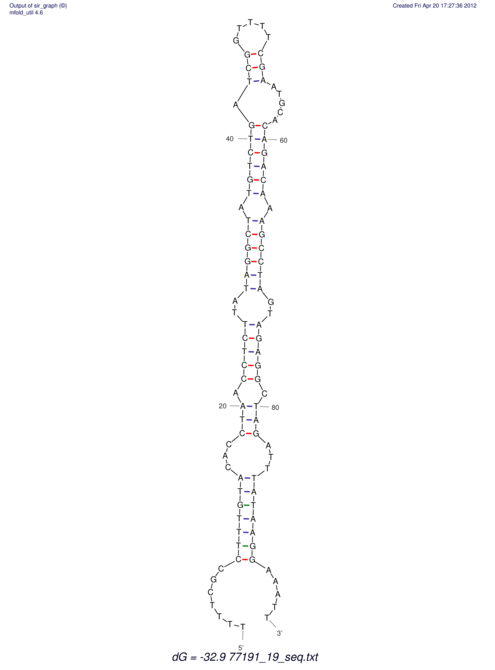
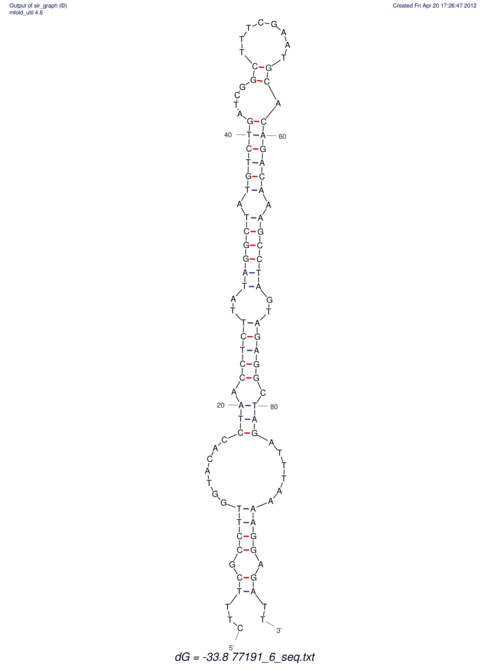
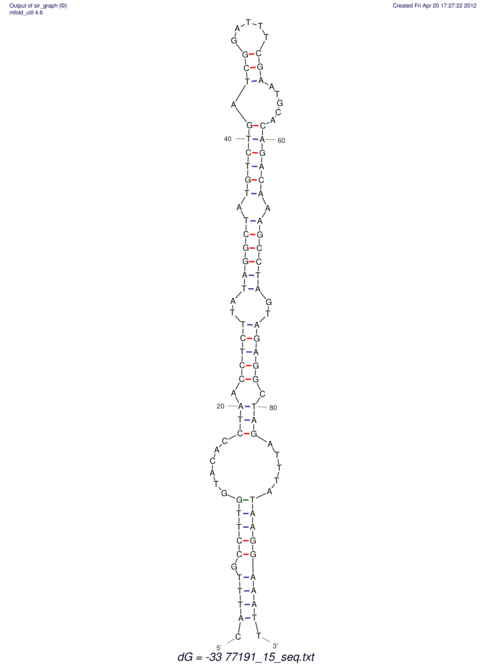
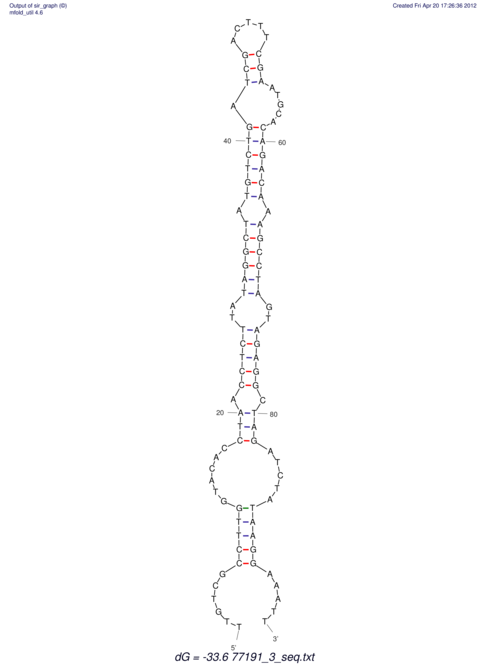
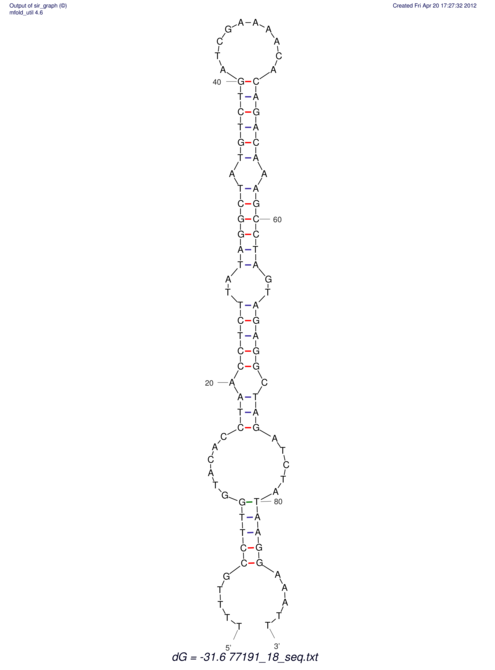
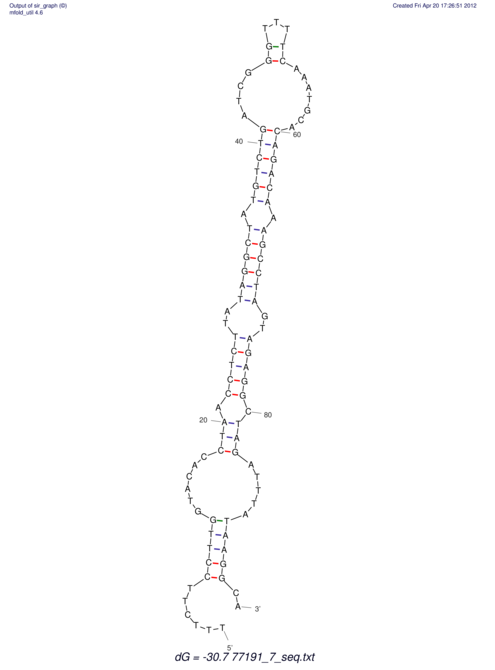
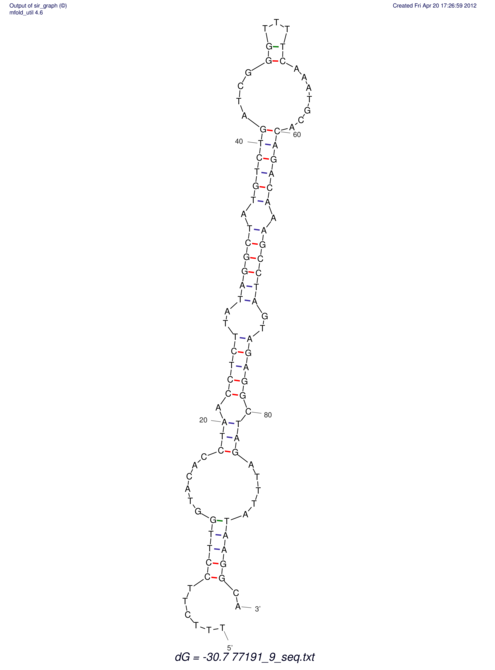
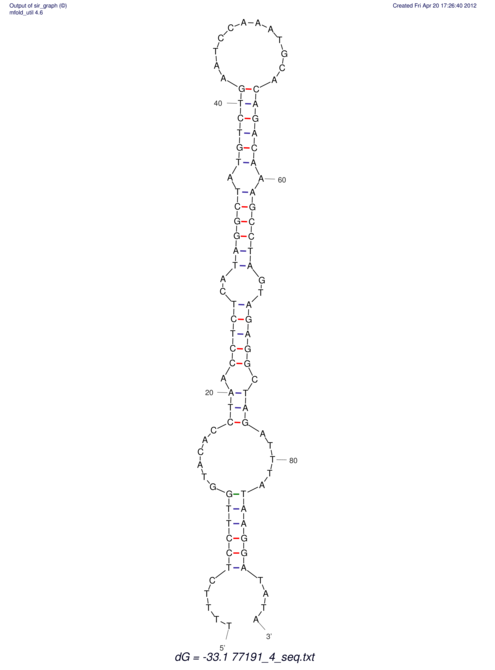
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:27:39 EDT