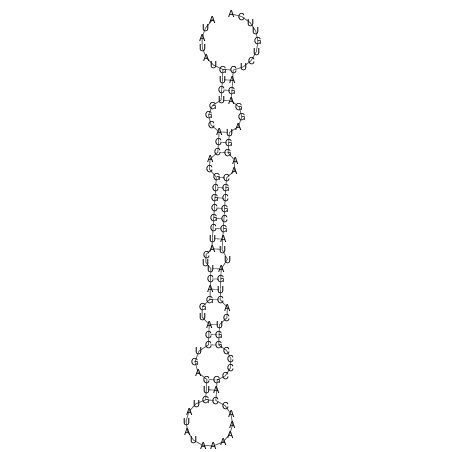
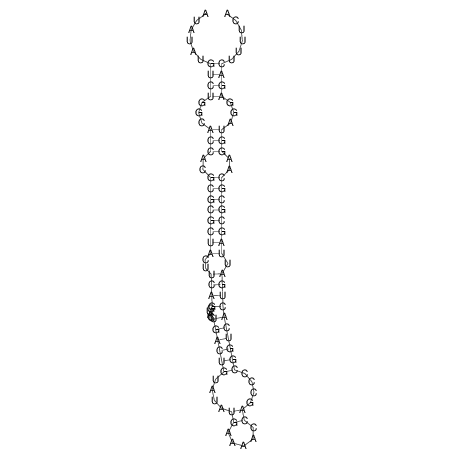
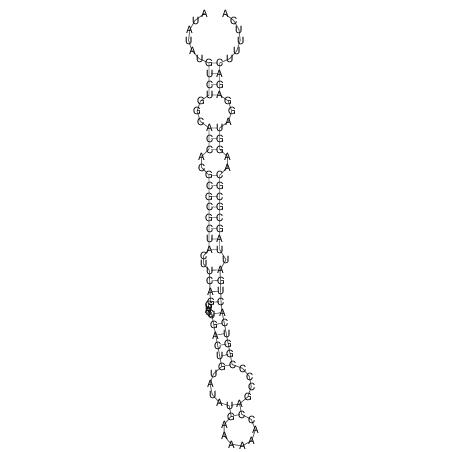
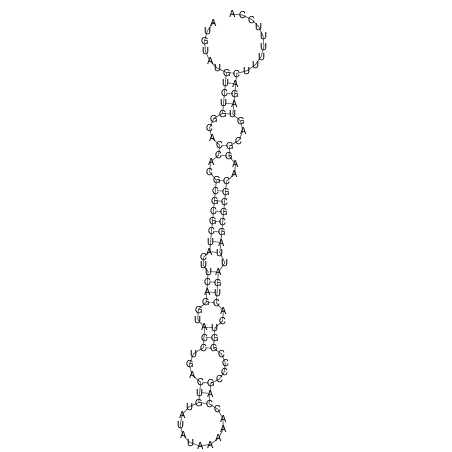
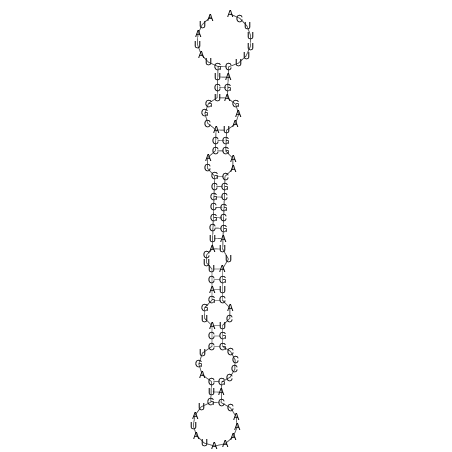
| dm3 |
chr2L:7425780-7425907 - |
NA |
ATGG--TGAA------------------GATGG-AG----ATA--TATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-AA-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTT-TA----------CAGTCGGG--GGAACA--T--CA |
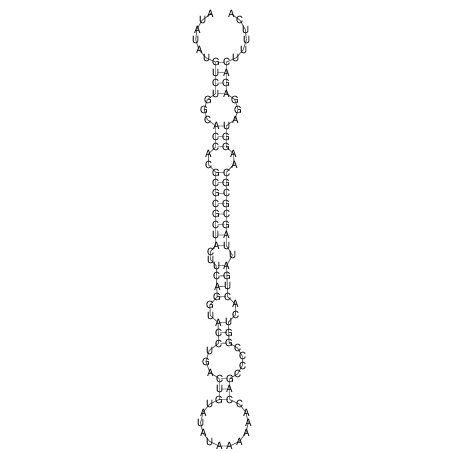
| droSim1 |
chr2L:7217350-7217477 - |
1 |
ATGG--CGGA------------------GATGG-AG----ATA--TATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-AA-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTT-TT----------CAGTCGGG--GGAACA--T--CA |
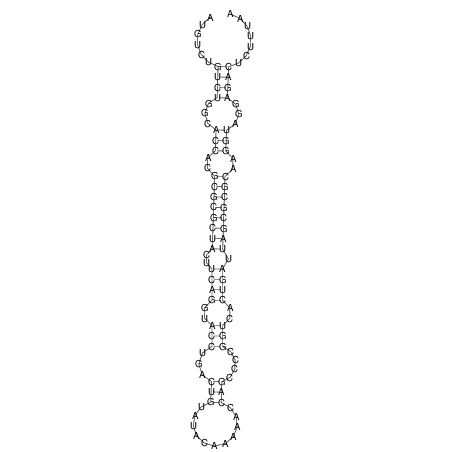
| droSec1 |
super_3:2941240-2941367 - |
1 |
ATGA--CGGA------------------GATGG-AG----ATA--TATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-AA-----AAAACAAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTT-TT----------CAGTCGGG--GGAACA--T--CA |
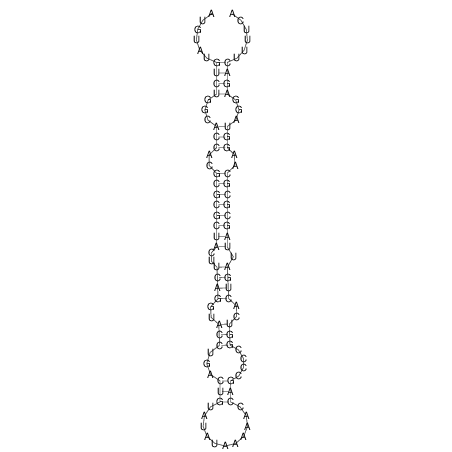
| droYak2 |
chr2L:5473766-5473897 + |
1 |
ATATCGTGGA------------------GTTGG-AG----ATA--TATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-AA-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTC-TG-------T-TCAGTCGGG--GGAACA--T--CA |
| droEre2 |
scaffold_4929:16344506-16344633 - |
1 |
ATGG--TGGA------------------GATGG-AG----ATA--TATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-AA-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTT-TT----------CAGTCGGG--GGAACA--T--CA |
| droEug1 |
scf7180000409463:614687-614823 - |
1 |
GCGA--TGAAG--------------ACATATGG-AG----ATG--TATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATGT--A-----AAAACCAGCCCCGGTCTCTGATTAGCGCGCAAGGTAGAAGACAT-TTAAGCCTTT-TCA---GGG--AGAACA--T--CA |
| droBia1 |
scf7180000302422:9125514-9125641 - |
1 |
GCGG--TGGA------------------TGTGG-AG----ATG--TCTGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAC--A-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTCTTT----------AAGCCGGG--GGAACA--T--CA |
| droTak1 |
scf7180000415604:142420-142542 + |
2 |
ATGG------------------------GTTGG-AG----ATG--TATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT--A-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTT-TT----------CAGCCGGG--GGAACA--T--CG |
| droEle1 |
scf7180000491028:1624837-1624954 + |
4 |
AAA--------------------------ATGA-AG----ATA--TATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT--G-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTT-TT----------CAGTTGG---GGAACA------- |
| droRho1 |
scf7180000766421:7648-7769 + |
2 |
------TGGA------------------AATGA-AG----ATA--TATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATATGAA-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTT-TT----------CAGCTGG---GGAACA------- |
| droFic1 |
scf7180000454115:155668-155793 - |
2 |
GTGC--TGGA------------------TATGG-AG----ATG--TATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT--A-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGCAGTAGACTT-TT-------TTCCAGC-TGG--GGAACA------- |
| droKik1 |
scf7180000302384:54817-54950 + |
1 |
GAGT--TGTT------------------G---GC-GATG-ATA--TATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT--A-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAAGAGACTTTTT----------CAGCTGGGAAGGAACACATCCCG |
| droAna3 |
scaffold_12916:15333988-15334092 - |
1 |
ATGG--CG---------------------------G----ATA--TATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGT-TG--CA-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTG-GA----------GAGT------------------- |
| droBip1 |
scf7180000396572:1689389-1689508 + |
1 |
ATGT--CGGA------------------TATGGCGG----ATA--TATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGT-TG--CA-----AAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTT-TG----------GAG---------TACA--T--GG |
| dp4 |
chr4_group1:4175727-4175841 - |
2 |
ACAC------------------------------AG----ATGTTCGTGTCTAG-CACCACGCGCGCTACTTCAGGTACCTGACTGCAGTG-AA-----AAAACCAGCCCCGGTCTCTGATTAGCGCGCATGGTAGGAGACTT-TC----------AAGAAGGT--G-----------G |
| droPer1 |
super_5:1086448-1086562 + |
1 |
ACAC------------------------------AG----ATGTTCGTGTCTAG-CACCACGCGCGCTACTTCAGGTACCTGACTGCAGTG-AA-----AAAACCAGCCCCGGTCTCTGATTAGCGCGCATGGTAGGAGACTT-TC----------AAGAAGGT--G-----------G |
| droWil1 |
scaffold_180772:5257564-5257692 + |
2 |
GTGT--CGTCGTTGTCGCCGTCGTC--GT---GC-GATGAATC--TTGGTCTAA-AACCACGCGCGCTACTTCAGGTACCTGACTGTATTG-AATAAAATTACTCAGCCCTGGTCACTGATTAGCGCGCAAGGTAAGAGAC-------------------------------------- |
| droVir3 |
scaffold_12963:15419953-15420066 + |
1 |
AAGC--TTAA-------------------------GATG-A-A--TACGTCTGGCCACCACGCGCGCTACTTCAGGTACCTGACTGTGG---GATAACCAAAACCAGCCCCGGTCACTGATTAGCGCGCACGGTAAAAGACGG-CG----------CAG-------------------- |
| droMoj3 |
scaffold_6500:18399385-18399491 + |
4 |
GCAG------------------------A---GC-GTTG-A-A--TGCGTCTAGCCACCACGCGCGCTACTTCAGGTACCTGACTGTGA---GATAATCGAAACCAGCCCCGGTCACTGATTAGCGCGCACGGTAAAAGAC-------------------------------------- |
| droGri2 |
scaffold_15126:5008401-5008495 - |
5 |
ATG--------------------------------------------CGTCTGGCCACCACGCGCGCTACTTCAGGTACCTGACTGTGG---GATAATCAAAACCAGCCCCGGTCACTGATTAGCGCGCGCGGTAAACGAC-------------------------------------- |