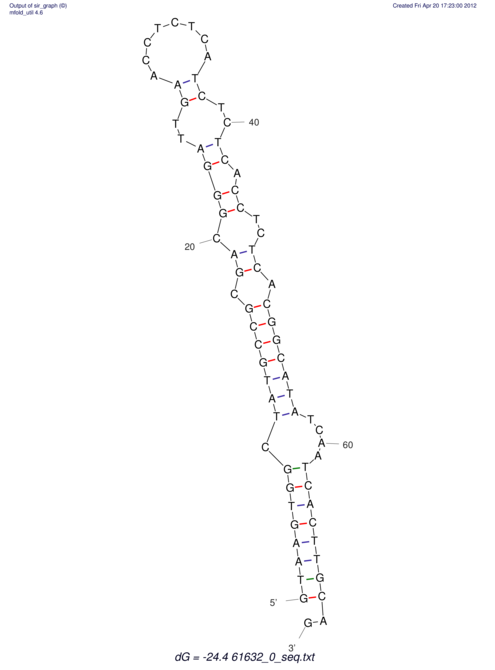
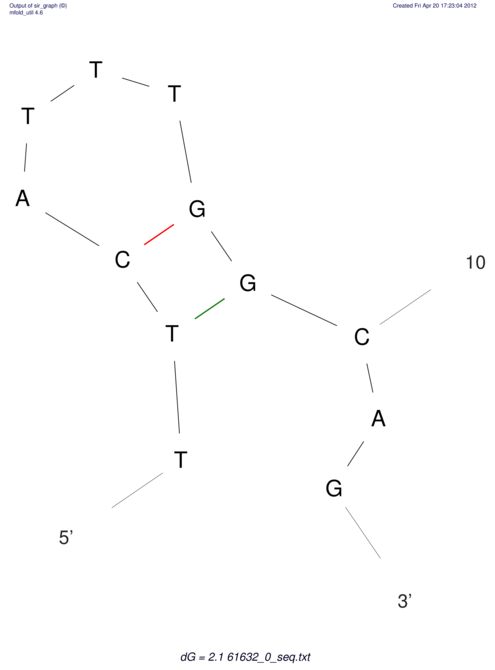
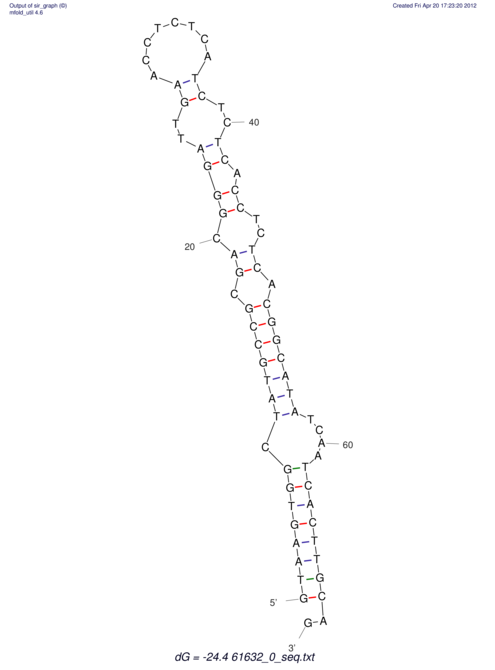
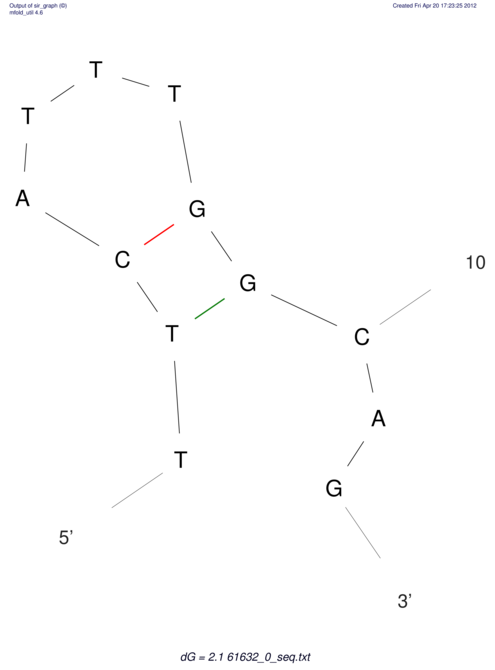
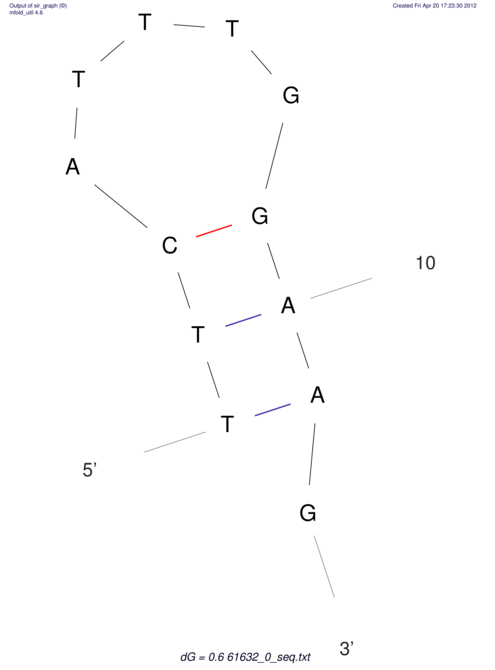
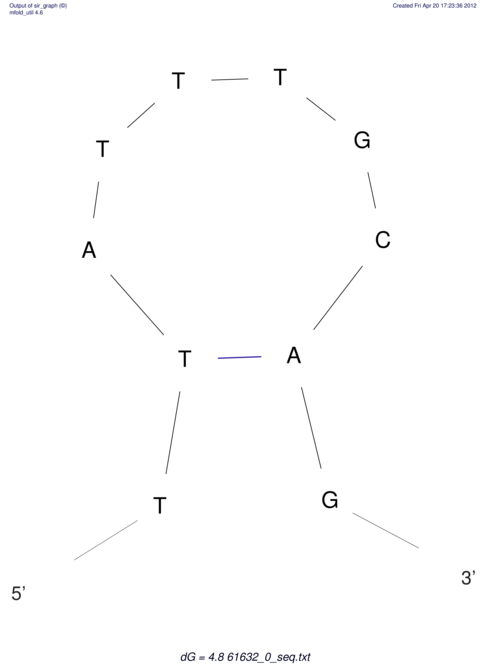
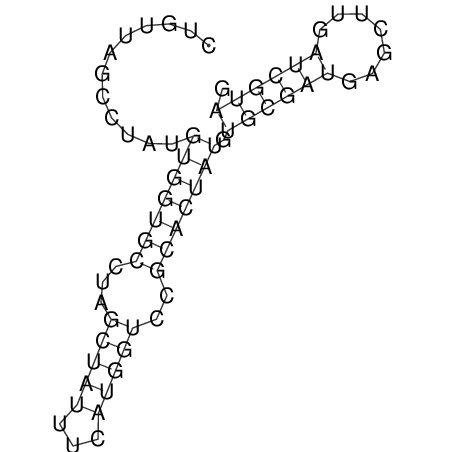
| ACGTACATCACTAAGGTAAGTGACTATGCCGGGATTGTGGTTATTGAACTTAGCGTCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG ACGTACATCACCAAGGTAAGTGACTATGCCGGGATTGTGGTTATTATACTTAGCGTCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| ............................................................ACTCACGGCATTTCATTCACTTG.................. | 23 | 1 | 358.00 | 0.00 | 294.00 | 39.00 | 25.00 |
| ............................................................ACTCACGGCATTTCATTCACTT................... | 22 | 1 | 89.00 | 3.00 | 79.00 | 5.00 | 2.00 |
| ............................................................ACTCACGGCATTTCATTCACT.................... | 21 | 1 | 81.00 | 3.00 | 73.00 | 2.00 | 3.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGCAG............... | 26 | 1 | 50.00 | 2.00 | 34.00 | 5.00 | 9.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGCA................ | 25 | 1 | 34.00 | 3.00 | 22.00 | 6.00 | 3.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGC................. | 24 | 1 | 34.00 | 0.00 | 29.00 | 5.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGGAT.................................................................. | 20 | 1 | 31.00 | 0.00 | 24.00 | 6.00 | 1.00 |
| ...............GTAAGTGACTATGCCGGGATTGTGG............................................................. | 25 | 1 | 29.00 | 1.00 | 6.00 | 4.00 | 18.00 |
| ...............GTAAGTGACTATGCCGGGATT................................................................. | 21 | 1 | 28.00 | 0.00 | 18.00 | 7.00 | 3.00 |
| ...............GTAAGTGACTATGCCGGGA................................................................... | 19 | 1 | 13.00 | 1.00 | 8.00 | 1.00 | 3.00 |
| ............................................................ACTCACGGCATTTCATTCAC..................... | 20 | 1 | 11.00 | 0.00 | 10.00 | 1.00 | 0.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGCAGT.............. | 27 | 1 | 8.00 | 5.00 | 1.00 | 0.00 | 2.00 |
| ............................................................ACTCACGGCATTTCATTCACTGG.................. | 23 | 1 | 6.00 | 0.00 | 4.00 | 1.00 | 1.00 |
| ............................................................ACTCACGGCATTTCATTCACC.................... | 21 | 1 | 6.00 | 0.00 | 5.00 | 1.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGGATTGT............................................................... | 23 | 1 | 4.00 | 0.00 | 2.00 | 1.00 | 1.00 |
| ...............GTAAGTGACTATGCCGGGATTG................................................................ | 22 | 1 | 4.00 | 0.00 | 2.00 | 0.00 | 2.00 |
| ................TAAGTGACTATGCCGGGATTGT............................................................... | 22 | 1 | 3.00 | 0.00 | 1.00 | 2.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGGATTGTG.............................................................. | 24 | 1 | 3.00 | 1.00 | 1.00 | 0.00 | 1.00 |
| ...............GTAAGTGACTATGCCGGGATTGTGGTT........................................................... | 27 | 1 | 3.00 | 0.00 | 0.00 | 2.00 | 1.00 |
| ............................................................ACTCACGGCATTTCATTCACG.................... | 21 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGGATTGTGT............................................................. | 25 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ........................................TTATTATACTTAGCGTCTCC......................................... | 20 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ..............................................................TCACGGCATTTCATTCACTTGC................. | 22 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ............................................................ACTCACGGCATTTCATTC....................... | 18 | 1 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGT................. | 24 | 1 | 2.00 | 0.00 | 0.00 | 1.00 | 1.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGCGG............... | 26 | 1 | 2.00 | 0.00 | 1.00 | 1.00 | 0.00 |
| ............................................................ACTCACGACATTTCATTCACTTG.................. | 23 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ...............................................................TACGGCATTTCATTCACTTG.................. | 20 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| .............................................................CTCACGGCATTTCATTCACTTG.................. | 22 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGA................. | 24 | 1 | 2.00 | 0.00 | 0.00 | 2.00 | 0.00 |
| ............................................................GCTCACGGCATTTCATTCACTTGCAG............... | 26 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ...............GTGAGTGACTATGCCGGGATTGTGG............................................................. | 25 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTGACTATGCCGGGATTGG............................................................... | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGCAGG.............. | 27 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ............................................................ACTCACGGCATTTCATTCACTTAA................. | 24 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGGATTTTGG............................................................. | 25 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ............................................................ACTCACGTCATTTCATTCACTTG.................. | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .............................................................CTCACGGCATTTCATTCACTTGCAGT.............. | 26 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ..............................................................TCACGGCATTTCATTCACTTGCAGT.............. | 25 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ............................................................ACTGACGGCATTTCATTCACTTG.................. | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ............................................................ACTCACGGGATTTCATTCACTTG.................. | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ............................................................ACTCACGGCATTTCATTCCCTTGCAGT.............. | 27 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ............................................................ACTCATGGCATTTCATTCACTT................... | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...............................................................CACGGCATTTCATTCACTTGCAG............... | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ........................TATGCCGGGATTGTGGTTATT........................................................ | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGGG................................................................... | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGCAC............... | 26 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGGATTGTGGT............................................................ | 26 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTGACTATGCCGGGAA.................................................................. | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .............................................................CTCACGGCATTTCATTCACTTGCT................ | 24 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .................AAGTGACTATGCCGGGAT.................................................................. | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ..............................................................TCACGGCATTTCATTCAC..................... | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGGATTGTGGAA........................................................... | 27 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTGACTATGCCGGGATTT................................................................ | 22 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ............................................................ACTCACGGCATTTCATTCACTTGCCG............... | 26 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ............................................................ACTCACGGAATTTCATTCACTTG.................. | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ........................................TTATTATACTTAGCGTCTCCACT...................................... | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ............................................................ACTCGCGGCATTTCATTCACTTGCAG............... | 26 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ................TAAGTGACTATGCCGGGATTG................................................................ | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGGATG................................................................. | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ............................................................ACTCACGGCATTTCTTTCACC.................... | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...............GTAAGTGACTATGCCGGCATTGTGG............................................................. | 25 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ..............................................................TCACGGCATTTCATTCACTTG.................. | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ................................................CTTAGCGTCTCCACTCACGGC................................ | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ACGTACATCACTAAGGTAAGTGACTATGCCGGGATTGTGGTTATTGAACTTAGCGTCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG ATGGACAACGGTAACGAAATTATTCTTGCAACCATTGACGTTATATTATCTAATGATTC-TCATACTCTATGTCGTCCA--TCGAAGAGATCGTCGAGGAA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| ACGTACATCACTAAGGTAAGTGACTATGCCGGGATTGTGGTTATTGAACTTAGCGTCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG GTGGTTATC----AGATTGACAACGGTAACGAAATTATAGCCGTGAAGGTTATATTATCTAATGATTCTCATACTCTATGTTGCAGTCGATCGTCGAGGAA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| ..................................................................................GCAGTCGATCTTCGAGGG. | 18 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ACGTACATCACTAAGGTAAGTGACTATGCCGGGATTGTGGTTATTGAACTTAGCGTCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG ACCATCCTAACTAGGGTTCGTGCAATTCCATAGCCAAACATTC--CAGCTTAC-A-TTCAACTTTCGC--TTTCCCACTTTTCCAGCTGATCGTTGAGGAA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| ACGTACATCACTAAGGTAAGTGACTATGCCGGGATTGTGGTTATTGAACTTAGCGTCTCCACTCACGGCATTTCATTCAC-----TT--GCAGTACATCACCGCCGAG ATCACAATTGTAAAGGTATG-------------------ATAAAAAAACTAAGTTCATTAATAAAACCCCCTTAACCTCATTTTGTTGCAAAGTATGTAGTAGCTGAA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| ACGTACATCACTAAGGTAAGTGACTATGCCGGGATTGTGGTTATTGAACTTAGCGTCTCCACTCACGGCATTTCATT--CACTTGCAGTACATCACCGCCGAG CCGT----CACCATATCCGCCGCATATCTTGCCCAAGTATGCACTGAGTTATTTACTTCCATATATGCTAGCTTAATGGTATTTTTAGATGATTGTGATGGAA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| ACGTACATCACTAAGGTAAGTGACTATGCCGGGATTGTGGTTATTGAACTTAGCGTCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG GCCTCGA---------------------------CTGTGACCA-----------------------------------------AGTATGTGACCAGCGAA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| .CCTCGA---------------------------CTGTxACCA-----------------------------------------AGTC............. | 18 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |