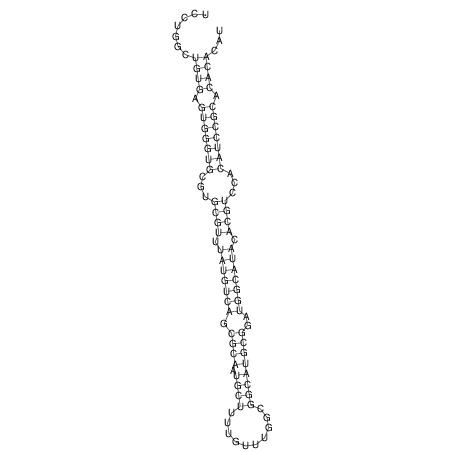
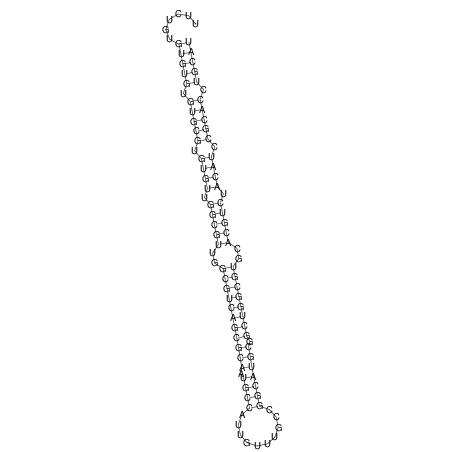
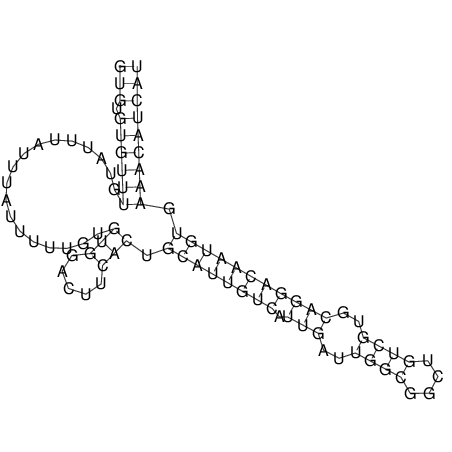
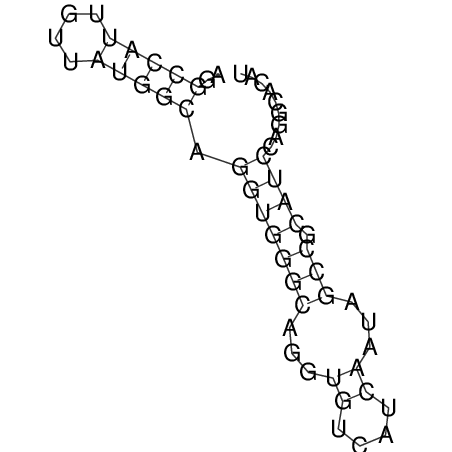
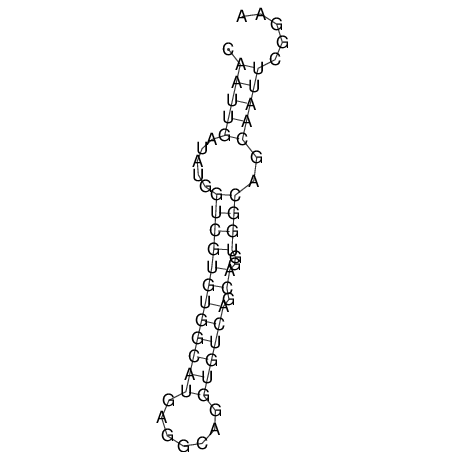
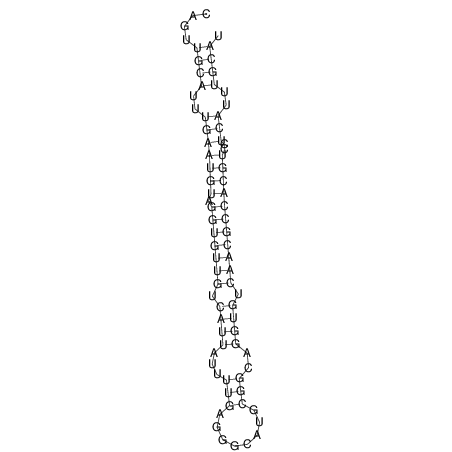
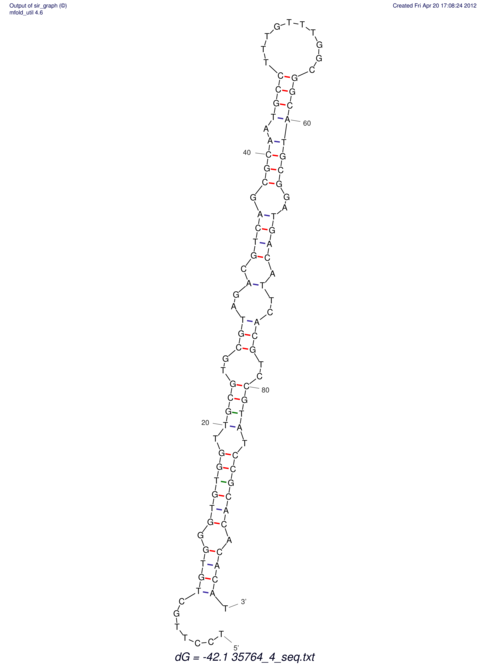
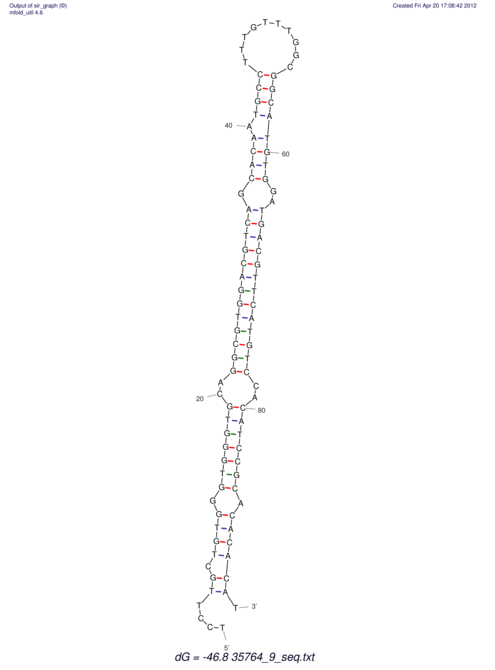
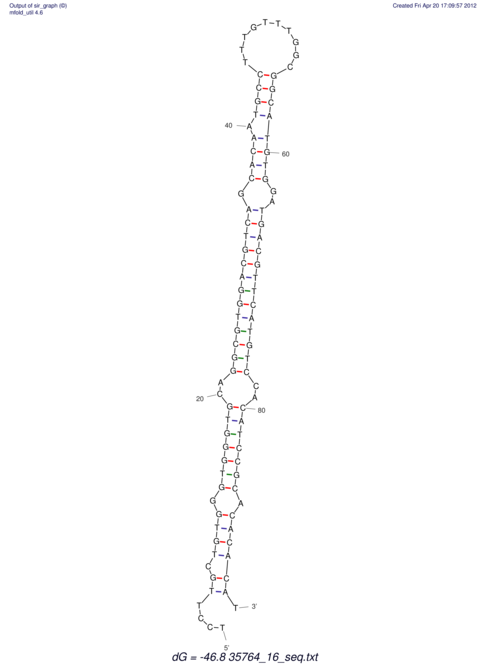
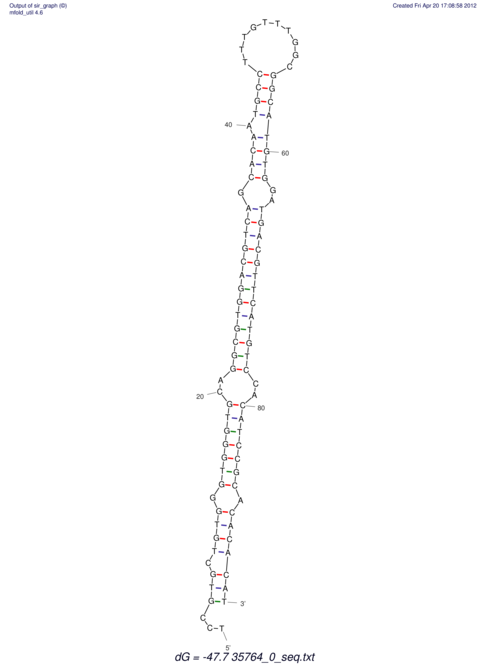
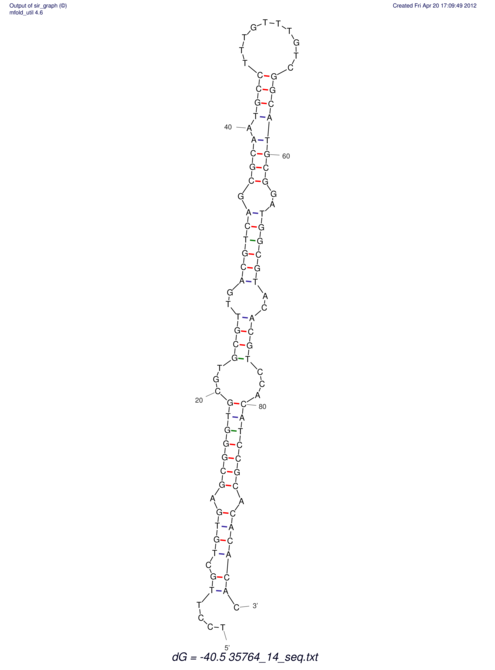
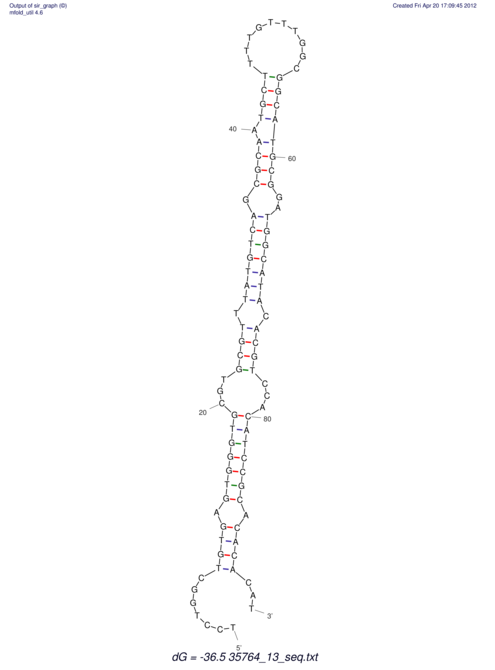
| CTTTGGGCGTTCGCCTCCGTGCTGTGGGT------GGGT--------GCAGGCGTGGACGTCAGCACAATGCCTTTGTTTGGCGGCAT----GTGGATG---ACGTTCATGTCCACATCCGCACACACATCC-----GCATCCA--TATC | Size | Hit Count | Total Norm | V113
Male body | V114
Embryo | V115
Head |
| ...............................................GCAGGCGTGGAGGTCAGT..................................................................................... | 18 | 1 | 247.00 | 86.00 | 89.00 | 72.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGTCCA................................... | 22 | 1 | 225.00 | 167.00 | 20.00 | 38.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGTCC.................................... | 21 | 1 | 134.00 | 124.00 | 4.00 | 6.00 |
| .....................................................................................CAT----GTGGATG---ACGTTCATGTCCA................................... | 23 | 1 | 128.00 | 61.00 | 36.00 | 31.00 |
| .......................GTGGGT------GGGT--------GGAGGCGA............................................................................................... | 18 | 1 | 121.00 | 0.00 | 0.00 | 121.00 |
| ......................................G--------GCAGGCGTGGAGGTCAG...................................................................................... | 18 | 1 | 32.00 | 22.00 | 6.00 | 4.00 |
| .................................................AGGCGTGGACGTCAGCACAATGC.............................................................................. | 23 | 1 | 22.00 | 14.00 | 5.00 | 3.00 |
| .................................................AGGCGTGGACGTCAGCACAATG............................................................................... | 22 | 1 | 18.00 | 13.00 | 2.00 | 3.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGTC..................................... | 20 | 1 | 17.00 | 17.00 | 0.00 | 0.00 |
| .....................................................................................CAT----GTGGATG---ACGTTCATGTCC.................................... | 22 | 1 | 17.00 | 7.00 | 3.00 | 7.00 |
| .................................................AGGCGTGGACGTCAGCACAATGCC............................................................................. | 24 | 1 | 14.00 | 8.00 | 4.00 | 2.00 |
| ......................................G--------GCAGGCGTGGAGGTCAGC..................................................................................... | 19 | 1 | 6.00 | 5.00 | 1.00 | 0.00 |
| .....................................................................................CAT----GTGGATG---ACGTTCATGTC..................................... | 21 | 1 | 5.00 | 3.00 | 2.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGT...................................... | 19 | 1 | 5.00 | 4.00 | 0.00 | 1.00 |
| .......................................................................................T----GTGGATG---ACGTTCATGTCCA................................... | 21 | 1 | 5.00 | 5.00 | 0.00 | 0.00 |
| .......................GTGGGT------GGGT--------GGAGGCGG............................................................................................... | 18 | 1 | 4.00 | 0.00 | 0.00 | 4.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGTCCAC.................................. | 23 | 1 | 4.00 | 2.00 | 1.00 | 1.00 |
| ......................CGTGGGT------GGGT--------GGAGGCG................................................................................................ | 18 | 1 | 3.00 | 0.00 | 0.00 | 3.00 |
| .....................................................................................CAT----GTGGATG---ACGTTCATGTCCAC.................................. | 24 | 1 | 3.00 | 2.00 | 0.00 | 1.00 |
| ......................................................................................AT----GGGGATG---ACGTTCATGTCCA................................... | 22 | 1 | 3.00 | 3.00 | 0.00 | 0.00 |
| .................................................AGGCGTGGACGTCAGCACAAT................................................................................ | 21 | 1 | 2.00 | 2.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGTCCT................................... | 22 | 1 | 2.00 | 1.00 | 0.00 | 1.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGTCT.................................... | 21 | 1 | 2.00 | 2.00 | 0.00 | 0.00 |
| .....................................................................................CAT----GTGGATG---ACGTTCATTTCCA................................... | 23 | 1 | 2.00 | 2.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGACC.................................... | 21 | 1 | 2.00 | 2.00 | 0.00 | 0.00 |
| ....................................................................................GCAT----GTGGATG---ACxTTCAAGT...................................... | 20 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ...............................................GCAGGCGTGGAGGTCAGGA.................................................................................... | 19 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ................................................CAGGCGTGGAGGTCAGTA.................................................................................... | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ......................................................................................AT----GTGGATG---ACGTTCCTGTCC.................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGCTCATGTCCA................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GGGGATG---ACGGTCATGTCCA................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .......................................................................................A----GTGGATG---ACGTTCATGTCCA................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ...............................................GCAGGCGTGGATGTCAGT..................................................................................... | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ......................................................................................AT----GTGGCTG---ACGTTCATGTCCA................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---AAGTTCATGTCC.................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .......................................................................................T----GTGGATG---ACGTTCATGTC..................................... | 19 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .....................................................................................CAT----GTGGATG---ACGTTCGTGTCCAC.................................. | 24 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ......................................................................................AT----GTGGATG---CCGTTCATGTCC.................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................T--------GCAGGCGTGGAGGTCAGT..................................................................................... | 19 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTGCATGTCCA................................... | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .....................................GG--------GCAGGCGTGGAGGTCA....................................................................................... | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATCTCC.................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----ATGGATG---ACGTTCATGCCCA................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .....................................................................................CAT----GTGGATG---GCGTTCATGTCCA................................... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .....................................................................................CAT----GTGGATG---ACGGGCATG....................................... | 19 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .....................................AG--------GCAGGCGTGGACGTCA....................................................................................... | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGCCCA................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGTCCC................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGGCCA................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCACGTCC.................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGAC..................................... | 20 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCGTGTC..................................... | 20 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..TTGCGCxTTCGCCTCCGTGC................................................................................................................................ | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ......................................................................................AT----GTGGATG---ACGCTCATGTCCAC.................................. | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GAGGATG---ACGTTCATGTCCAC.................................. | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGTCCATT................................. | 24 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGCTCATGTCC.................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AT----GTCGATG---ACGTTCATGTCCAC.................................. | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .................................................AGGCGTGGACGTTAGCACAATG............................................................................... | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATA....................................... | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .................................................AGGCGTGGACGTAAGCAAAATG............................................................................... | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ......................................................................................AT----GTGGATG---ACGTTCATGACCA................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................................................................................AA----GTGGATG---ACGTTCATGTCCA................................... | 22 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .................................................AGGCGTGGACGTCAGCCCAATGC.............................................................................. | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ....................................AGT--------GCAGGCGTGGAGGTC........................................................................................ | 18 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .....................................................................................CAT----GTGGATG---ACGTTCATGTCCAA.................................. | 24 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ......................................................................................AT----GTGGACG---ACGTTCATGTCCA................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..................................................GGCGTGGACGTCAGCACAATG............................................................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |