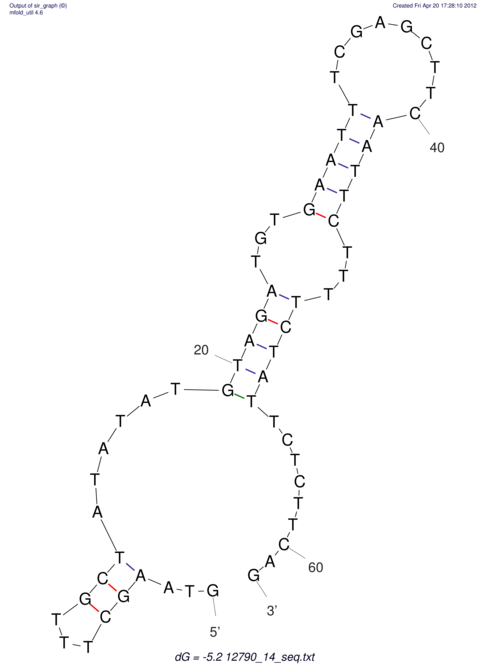
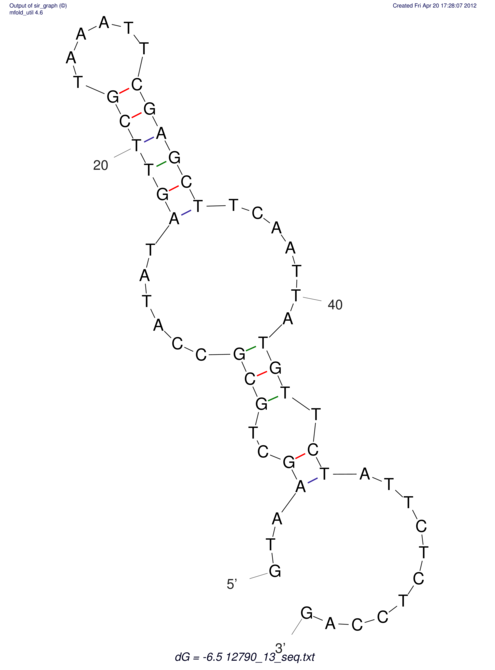
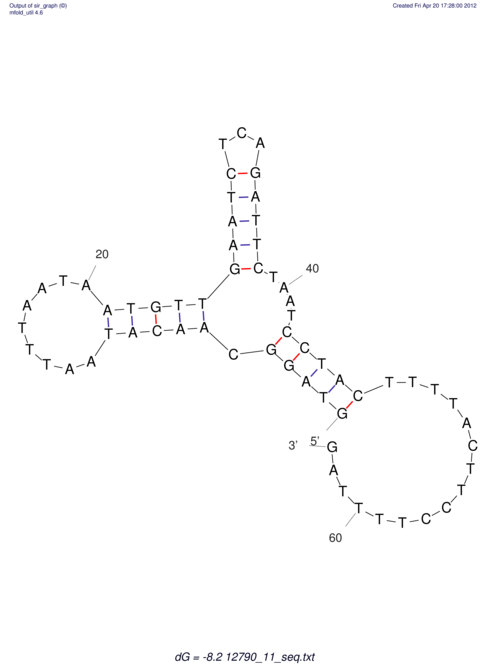
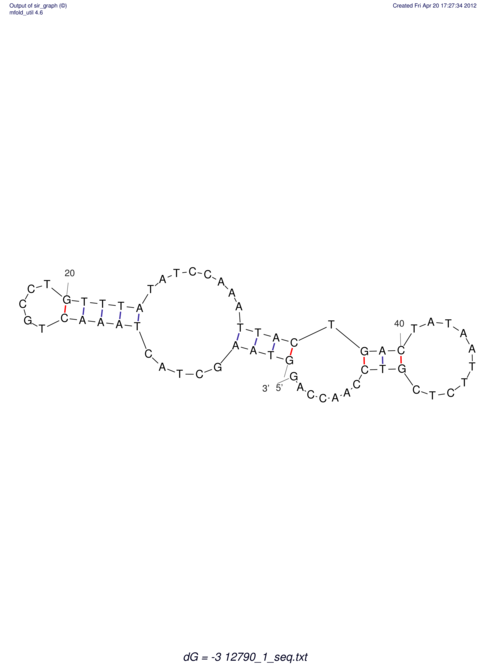
| dm3 |
chr2L:16678235-16678332 + |
|
1 |
GATAAGCACGTCATCGTAAGCATTGCAAA--------------------------------------------ATG-----T-------------AG------------------TT------------------------------------------------------------------GTTAAATTCGCTCTTCAATCATTAACA-ACA-TTTACAATGT--TTGCAGGCTGTCAACGTTGAT |
| droSim1 |
chr2L:16366233-16366322 + |
LASTZ |
5 |
GATAAGCACGTCATCGTAAGCATTGCAAA--------------------------------------------ATA-----T-------------AG------------------TT------------------------------------------------------------------GTTAAATTCGCTCTTCAATTATTTA-------C---TATTGA--TTGCAGGCTGTCAACGTTGAT |
| droSec1 |
super_7:325110-325199 + |
LASTZ |
5 |
GATAAGCATGTTATCGTAAGCATTGCAAA--------------------------------------------ATA-----T-------------AG------------------TT------------------------------------------------------------------CTTAAATTCGCTCTTCAATTATTTA-------C---TATTGA--TTGCAGGCTGTCAACGTTGAT |
| droYak2 |
chr2R:4424626-4424717 - |
LASTZ |
5 |
GATAAGCACGTCATCGTAAGCTTTGCTAT--------------------------------------------ATA-----T-----------GTAG------------------AT------------------------------------------------------------------GTGAATTTCGAGCTTCAATTCTTTT-------C---TATTCT--CTTCAGGCTGTCAATGTTAAT |
| droEre2 |
scaffold_4845:19748460-19748547 + |
LASTZ |
5 |
GATAAGCACGTTATTGTAAGCTGCGCCA----------------------------------------------TA-----T-------------AG------------------TT------------------------------------------------------------------CGTAAATTCGAGCTTCAATTATGTT-------C---TATTCT--CTCCAGGCTGTCAATGTTAAT |
| droEug1 |
scf7180000408961:253912-254004 + |
LASTZ |
5 |
AATAAGCAACTTATTGTAGGCAACATAAT--------------------------------------------TTA-----A-------------TA------------------AT----------------------GT-------------------------------------------TGAATCTCAGATTCTAATCCTACT-------TTT-ACTTCC--TTTTAGGCAGTCAATATGGAT |
| droBia1 |
scf7180000301468:836914-837003 + |
LASTZ |
5 |
GATAAGCAAGTCATTGTAAGCTACTAAAC--------------------------------------------T--GCCTGT---TTAT----AT-------------------------------------CCAAATTACT---------------------------------------------------------GACTATA---------ATTCTCGTCC--AACCAGGCTGTCAATGTAAAT |
| droTak1 |
scf7180000415914:402900-403000 + |
LASTZ |
5 |
GATAAGCAAGTTATTGTAAGCTACACATT--------------------------------------------TTA--TCTT---TTGCAACCAA-------------------------------------AGAG--AGCC------------------------------------------TA-TTCTAATAC-----------T--CTATTC---CACGCAACCCACAGGCAGTCAATGTGAAT |
| droEle1 |
scf7180000490579:668430-668616 - |
LASTZ |
5 |
AATAAACAAGTTATTGTGAGTCCAAAT--TTAAGGCTTTTTTAAGTACGAAAACTATCATCA-TTAATCCGTTAC----------ATTT--------------------------TTGACATGGGCTAGCTTTAACTAGGCTGAAAATTGAATATTATTACGATTACTAATATATATACTAATAC--------------GAACTTACT--TTG-TC---TTTTCT--CCTCAGGCGGTCAACGTGAAC |
| droRho1 |
scf7180000777347:13383-13513 - |
LASTZ |
5 |
AATAAACAAGTTATTGTAAGTAAGGAATT--AAGGCTTCTTTGAGTAA-----------TCA-T--AT-----TCG-----A-------------CAAGAGCAATTCACATATTTTTGACATGTTTTG----------------------------------------------------------------ACCT---ACTTCATCG------CT---ATATTT--TTCCAGGCTGTCAACGTTAAT |
| droFic1 |
scf7180000449814:1574-1709 - |
LASTZ |
5 |
AACAAACAAGTCATCGTAAGTAATCAAGG------CTTTCTTCATCAGAAAGGTTATTTTTAGCCAAA-----ATA-----CAGGATTT----TT-------------------------------------GGATTAAACTAA-----TAA--------------------------------TA-TTTTTACAT-----------T--TTGTACT--GCTTGT--CCCCAGGCTGTTAATGTAAAT |
| droKik1 |
scf7180000302382:508886-508975 + |
LASTZ |
5 |
GACAAGCAAGTGATTGTAAGTTAAGCGAA--------------------------------------------TTC-----A-------------AG------------------AT------------------------------------------------------------------TTTCTAGCCCTAGTACGACCCTTCT-------C---GTTTCT--GACCAGGCTGTCAACGTGAAT |
| droAna3 |
scaffold_12916:15881689-15881781 - |
LASTZ |
5 |
GAAAAGCAAGTGATTGTAAGATGTAAACTCTTAGACCCTCCTAACTG----------------------------------------------CT-------------------------------------ACA---AGCT---------------------------------------------------------AATACTTTT-------G---CTTTGA--ATCCAGGCTGTTAACGTGAAC |
| droBip1 |
scf7180000396717:120661-120753 - |
LASTZ |
5 |
GAAAAGCAAGTGATTGTAAGTTTCGAACTCTAAGACCCAAGTAGGTG----------------------------------------------CC-------------------------------------ACAAACTATA---------------------------------------------------------AGT---TTT-------G---CTTTCA--ATCCAGGCTGTTAACGTGAAC |
| dp4 |
chr4_group3:8422191-8422279 + |
LASTZ |
5 |
CAAAAGCAGCTGATAGTGAGTACTGGAAG--------------------------------------------C--ATTTAT---ATCT----CT-------------------------------------GGAAAGCACT---------------------------------------------------------AATTCTTTT-------T---TTAATC--TTCTAGGCTGTCGAATTAAAT |
| droPer1 |
super_1:9873274-9873362 + |
LASTZ |
5 |
CAAAAGCAGCTGATAGTGAGTACTGGAAG--------------------------------------------C--ATTTAT---ATCT----CT-------------------------------------GGAAAGCACT---------------------------------------------------------AATTCTTTT-------T---TGAATC--TTCTAGGCTGTCGAATTAAAT |
| droWil1 |
scaffold_181132:892502-892596 - |
MAF |
5 |
GACAAAGAAGTGATTGTAGGTGTCAATGA-------T------------------------------------T--------------T----CA-------------------------------------AACTAACATC--------AA------------------------------TTTTTTATTAAC-----GTCTTTATT-------T---TTTCCT--ATTCAGGCTATCAATGTCAAT |
| droVir3 |
Unknown |
MAF |
0 |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
Unknown |
MAF |
0 |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
|
0 |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |