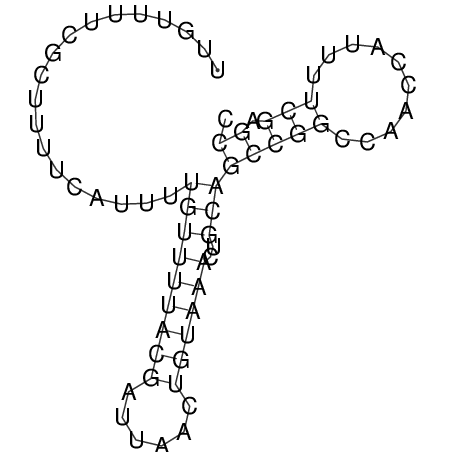
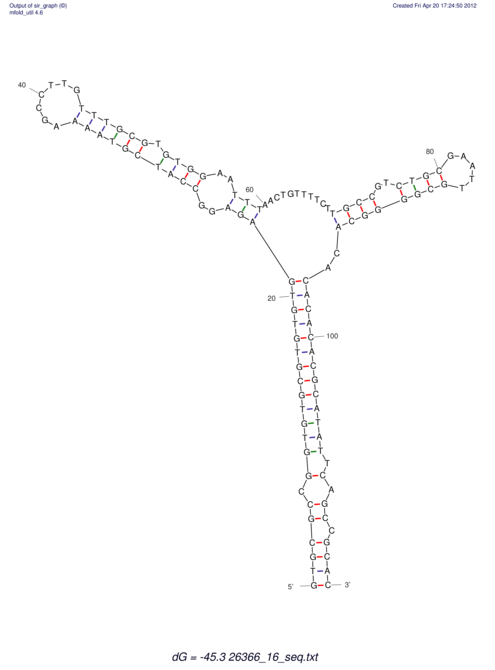
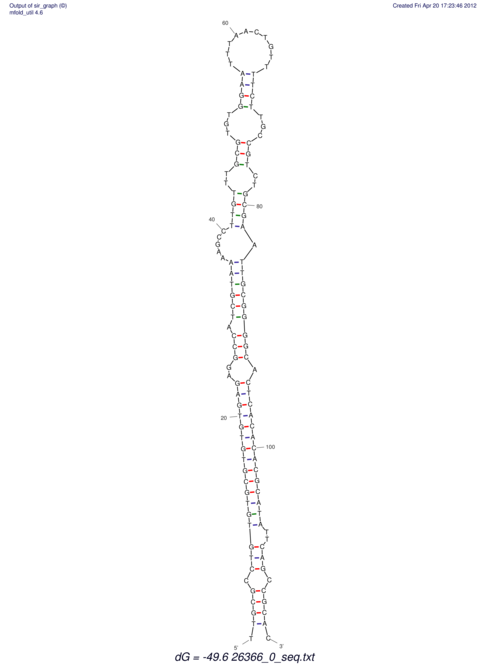
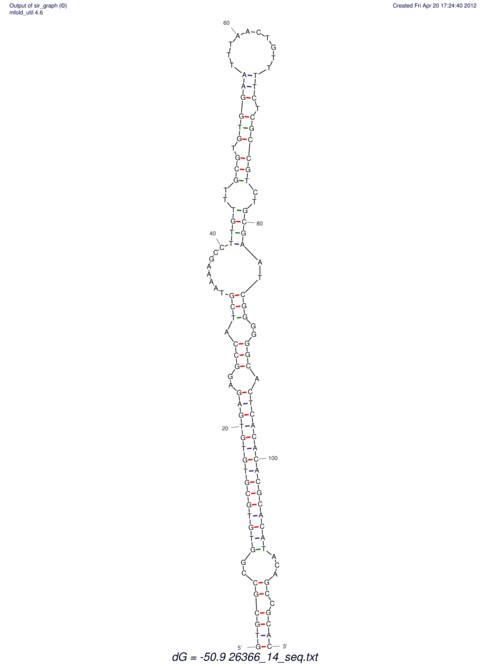
| dm3 |
chrX:1904318-1904465 + |
|
1 |
AAGAGC-GC------AAAAT-------------G-------------------AGTGCG--CCGGTG--TGCGTGT--G--T-------GAG----AGGCC--ATCGTAAAAGCCTTGTTTGCGTGCGGAATTTAACTGTTTTCTCGCCGTCTGCG--AATTGCG-------GGG--------CACTCACACA----------------CGCATATTCAGCCGCACATCTGGGAAAA------------------------TGGG |
| droSim1 |
chrX:1343126-1343273 + |
LASTZ |
5 |
AAGAGC-GC------GAAAT-------------G-------------------AGTGCG--CCGGTG--TGCGTGT--G--T-------GAG----AGGCC--ATCGTAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTCTTGCCGTCTGCG--AATTGCG-------GGG--------CACACACACA----------------CGCATATTCAGCCGCACATCTGGGAAAA------------------------TGGG |
| droSec1 |
super_10:1689518-1689665 + |
LASTZ |
4 |
AAGAGC-GC------AAAAT-------------G-------------------ATTGCG--CCTGTG--TGCGTGT--G--T-------GAG----AGGCC--ATCGTAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTCTTGCCGTCTGCG--AATTGCG-------GGG--------CACTCACACA----------------CGCATATTCAGCCGCACATCTGGGAAAA------------------------TGGG |
| droYak2 |
chrX:6422544-6422691 - |
LASTZ |
4 |
AAGAGC-GC------AAAAT-------------G-------------------TGTGCG--CCGGTG--TGCGTGT--G--T-------GAG----AGGCC--ATCGTAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTCTCGCCGTCTGCG--AATCGGG-------GGG--------CACTCACACA----------------CGCACATACAGCCGCACATCTGGGAAAA------------------------TGGG |
| droEre2 |
scaffold_4644:1874871-1875018 + |
LASTZ |
4 |
AAGAGC-GC------AAAAT-------------G-------------------TGTGCG--CCGGTG--TGCGTGT--G--T-------GAG----AGGCC--ATCGTAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTCTCGCCGTCTGCG--AATTGGG-------GGG--------CACTTACACA----------------CGCACATACAGCCGCACATCTGGGAAAA------------------------TGGG |
| droEug1 |
scf7180000409182:447470-447608 + |
LASTZ |
5 |
AACAGC-GA------AAAAT-------------G-T---TT-------GCGT------------ATG--TG----T--G--T-------GAG----GGGCC--A---TAAAAGCCTTGTTTGCGCGTGGAATTTAACTGTTTTTTCGCCGTCTGCT--GGTTCGG-------GGGGTGGGGAGCACGTACA------------------------CTCAACCGCACATCTGGGAAAA------------------------TGGG |
| droBia1 |
scf7180000301760:4508970-4509126 + |
LASTZ |
5 |
GACAGC-GC------AAATT-------------G-------------------TGTGCG--CTCGTGTGTGCGTGTG----TGT-----GTGTGCGGGGCC--G---TAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTCTCGCCGTCTGCC--AATTGGG-------GGG--------CACTCACACAG------------AGGCACACGCACAGTCACACACCTGGGAAAA------------------------TGGG |
| droTak1 |
scf7180000415211:36485-36643 - |
LASTZ |
5 |
GACGGC-GC------AAA-----------------T---GTGCCATGTGTGTGTGTGCG--CTGGTG--TGCGTGT--G--C-------G------GGGCC--G---CAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTCTCGCCGTCTGCG--AATTGGG-------GGG--------CACTCACACAG------------ACGCGCACACACAGCCGCACACCTGGGAAAA------------------------TGGG |
| droEle1 |
scf7180000491001:1416804-1416930 + |
LASTZ |
5 |
GAGAGC-GC------AAAAA-------------A-T---GT-------GGCT------------GTG--TG-------------------TG----TGGCC--A---TAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTCTCGCCGTCTGCA--AATTGGG-------GGG--------CACTAACACA------------------------CAGCCGTACACCTGGGAAAA------------------------TGGG |
| droRho1 |
scf7180000777610:12722-12848 - |
LASTZ |
5 |
AAGAGC-GC------AAAAA-------------A-T---GT-------GGCT------------GTG--TG-------------------TG----TGGCC--A---TAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTCTCGCCGTCTGCG--AATTGGG-------GGG--------CACTCACACA------------------------CAGCCACACACCTGGGAAAA------------------------TGGG |
| droFic1 |
scf7180000454072:3017522-3017650 + |
LASTZ |
5 |
GAGAGC-GC------AAAAT-------------G------G-------GACT--GT--------GTG--TGCGTGT---------------G----TGGCC--A---AAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTTTCGCCGTCTGCG--AATTGGG-------GGG--------AACACACA--------------------------CAGCTGCACACCTGGGAAAA------------------------TGGG |
| droKik1 |
scf7180000302610:169951-170123 + |
LASTZ |
5 |
AAAGGCGGAGAGCGCCGCAG-------------A-T---GT-------GCCA------------GTG--TGTGAGT--GTGTCTGTGTTGAG----TGGCC--A---TAAAAGCCTTGTTTGCGTGTGGAATTTAACTGTTTTCTCGCCGTCTGCG--AATTGGG-------GGG--------CACACACACAACCACACCAACCTA--CACGCACACACCGGCGCACCTGGGAAAA------------------------TGGG |
| droAna3 |
scaffold_12929:1897894-1898023 + |
LASTZ |
5 |
AAGGGG-GA------GAGAG-------------C---------AGTTTGCAT------------GTG--TGCGATT--T-----------TG----TGGTC--A---TAAAAGCTTTGTTTGCGTGTGGAATTTAACTGTTTTCTAGCCGTCTGCG--AATTGGG-------GGG--------GGGGG-----------------------------GGGCCATACAAGCTGGAAAA------------------------TAGG |
| droBip1 |
scf7180000396543:357925-358040 + |
LASTZ |
5 |
GGGGGA-AT----------A-------------G-T---TT-------GCAT------------GTG--TGCGATT--T-----------TG----TGGTC--A---TAAAAGCTTTGTTTGCGTGTGGAATTTAACTGTTTTCTAGCCGTCTGCG--AATTGGG-------GGG--------C-------------------------------------CATACAAGCTGGAAAA------------------------TAGG |
| dp4 |
chrXL_group1a:2576130-2576311 + |
LASTZ |
0 |
AAGAGG-GA------GAGATAGAGAGAGAGAATAATGGAG---TGCTGGTGTATATGTACCTACATATGTACGTGTGGT--T-------GAG----TACCC--A---TAAAAGCCTTGTTTGCGTGCTAGTTTTAACTGTTTTTTCGGTGTTCTTG--TGTTGGG-------CGG--------CACACACACC----------------AGCGCCATTGGCTTTTCACAGGGCGA----------------------GAAGTAAG |
| droPer1 |
super_28:1013167-1013344 + |
LASTZ |
0 |
AAGAGG-GA------GAGATAGAGAGAGAGAATAATGGAG---TGCTGGTGTATATGTA--CCTATG--TACGTGTGGT--T-------GAG----TACCC--A---TAAAAGCCTTGTTTGCGTGCTAGTTTTAACTGTTTTTTCGGTGTTCTTG--TGTTGGG-------CGG--------CACACACACC----------------AGCGCCATTGGCTTTTCACAGGGCGA----------------------GAAGTAAG |
| droWil1 |
scaffold_181096:6994288-6994457 - |
LASTZ |
0 |
GAGGCC-AA------AAGAG---A---------C-------------------AGTGCG--ATAGAG--AGAGGTA-------------GAG----AGAACGTACA-GAGAGAGCTTGTTCTGGTTTGTAACTAA-TTTTCCCC--CTCAAATGTAAGCATTTCGAATGCATGTG--------TGTGTACGTG------------------------TAATCACACACGCACACAAATACCACGTCATTACGATTTCCACATGGC |
| droVir3 |
scaffold_12928:6459245-6459266 + |
LASTZ |
0 |
GC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTATTTCTGTGCAAG------------------------TGTG |
| droMoj3 |
Unknown |
|
0 |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_15203:11679615-11679698 + |
LASTZ |
0 |
-------------------------------------------------------------------------------------------------------------------TTGTTTTCGCTTTTCATTT---TGTTTTACGATTAACTGTA--AACTGCA-------GCC--------GGC-----CA----------------ACCATTTTCGA---GCCCACTGGCAA----------------------AGAATGGG |