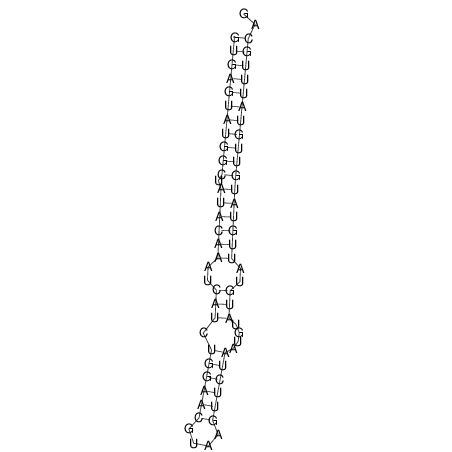
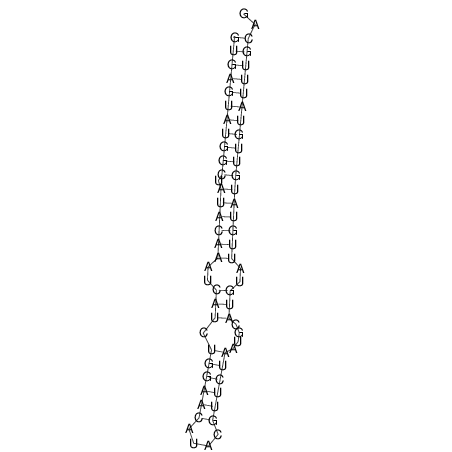
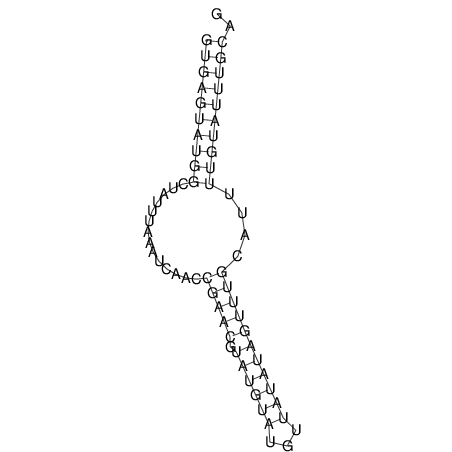
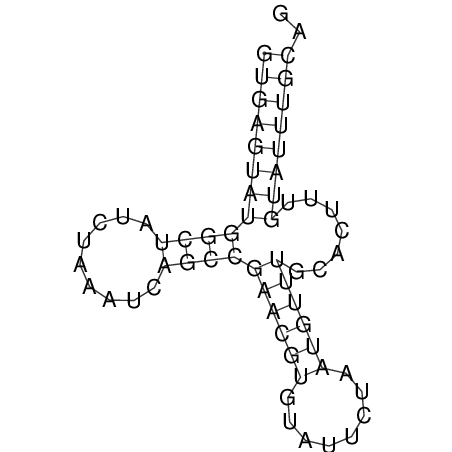
| AGCAATAAGTTAGACGTGAGTATGGC-------TATA-C----AAATCATCT-----G-GA---------ACATACG--T------TCTAATGCATGTA----TTGTA-TGT-TGTATTTGCAGTTTTATGATAGCAAA | Size | Hit Count | Total Norm | V113
Male body | V114
Embryo | V115
Head |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCAGT.............. | 22 | 1 | 131.00 | 13.00 | 100.00 | 18.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCAG............... | 21 | 1 | 100.00 | 9.00 | 19.00 | 72.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCA................ | 20 | 1 | 52.00 | 4.00 | 1.00 | 47.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCAGA.............. | 22 | 1 | 35.00 | 6.00 | 22.00 | 7.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGC................. | 19 | 1 | 10.00 | 0.00 | 0.00 | 10.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCAGC.............. | 22 | 1 | 5.00 | 0.00 | 3.00 | 2.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTCCAGT.............. | 22 | 1 | 2.00 | 0.00 | 2.00 | 0.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCTGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTCCAGA.............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .................................................................................................TA----TTGTA-TGT-CGTATTTGC................. | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TA----TTGTC-TGT-TGTATCTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TT----TTGTA-TGT-TGTATTTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTGAA-TGT-TGTATTTGCA................ | 20 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TA----TATTA-TGT-TGTATTTGCAG............... | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ...........................................AAATCATCT-----G-GA---------ACATACG--T------T.................................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .................................................................................................GA----TTGTA-TGT-TGTATTTGC................. | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ..................................................................................................A----TTGTA-TGT-TGTATTTGC................. | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................CTA----TTGTA-TGT-TGTATTTGCAGC.............. | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTCTA-TGT-TGTATTTGCAG............... | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCAT............... | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................CA----TTGTA-TGT-TGTATTTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ..................................................................................................A----TTGTA-TGT-TGTATTTGCAG............... | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTGTA-TTT-TGTATTTGCAG............... | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TA----TTGTA-TGT-TCTATTTGCA................ | 20 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .......................................................................................................TTGTA-TGT-TGTATTTGCA................ | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCCG............... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCAGTA............. | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTGTA-TTT-TGTATTTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTGTA-CGT-TGTATTTGCAGA.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCCGA.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCAGAT............. | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .................................................................................................GA----TTGTA-TGT-TGTATTTGCA................ | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTGTA-TGT-TGTATTTGCAGG.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................TA----TTGTA-CGT-TGTATTTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |