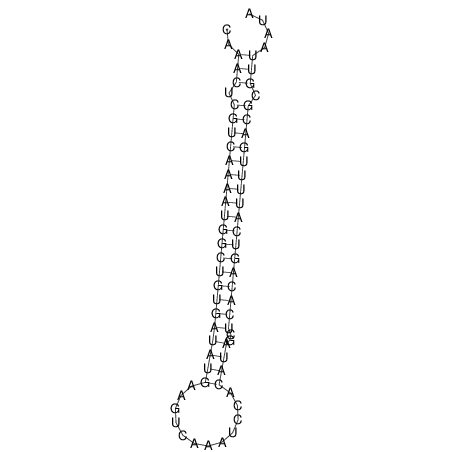
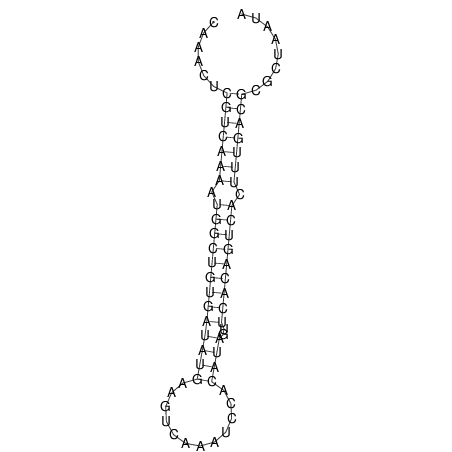
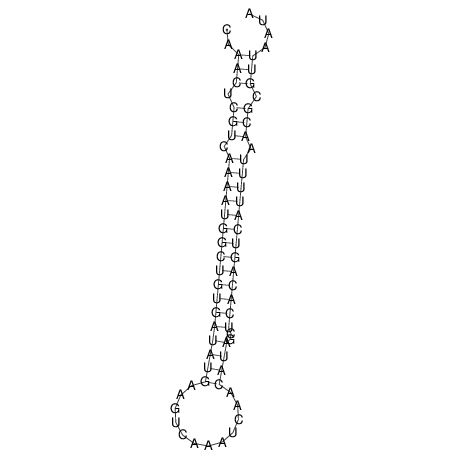
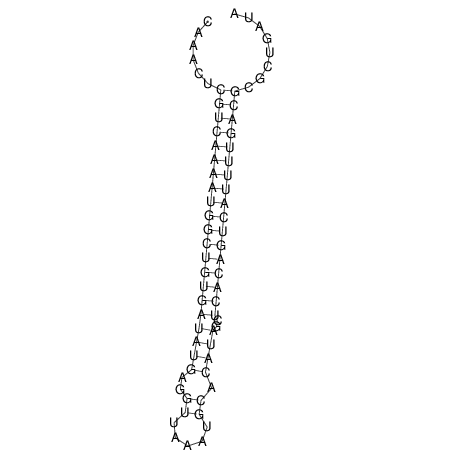
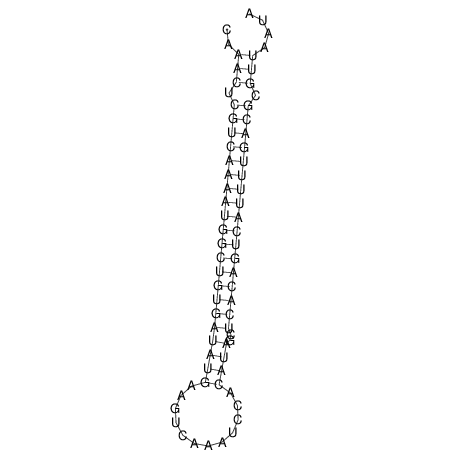
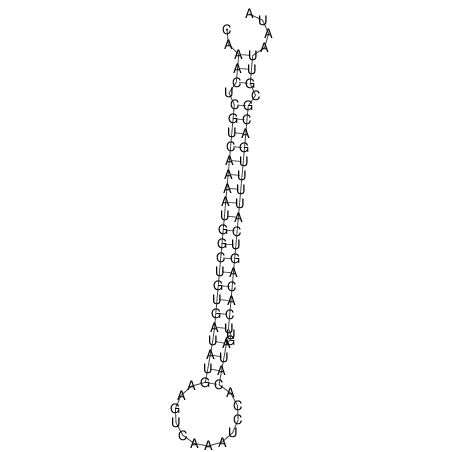
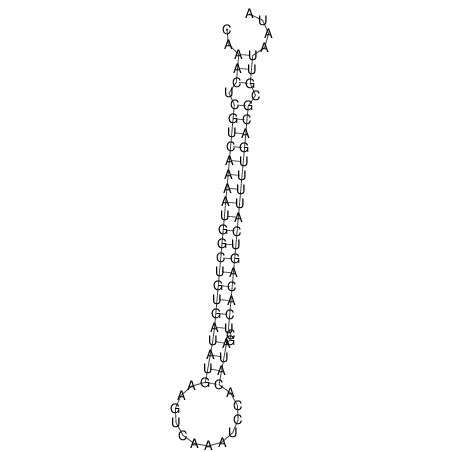
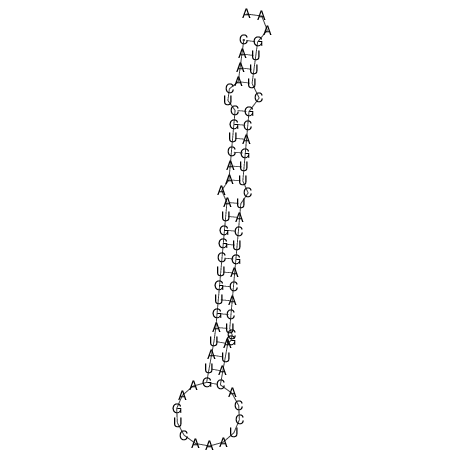
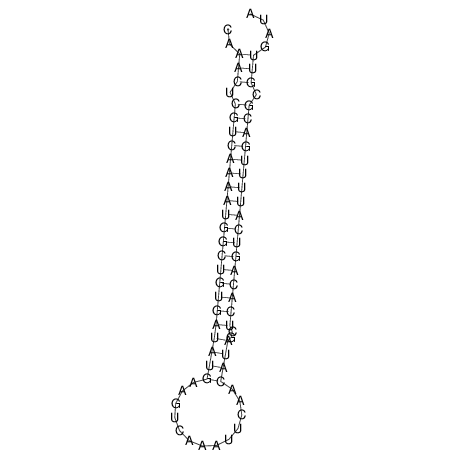
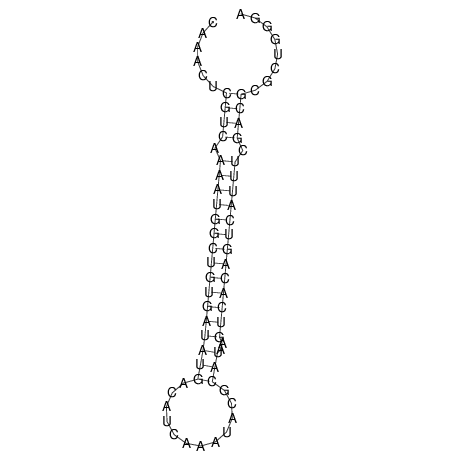
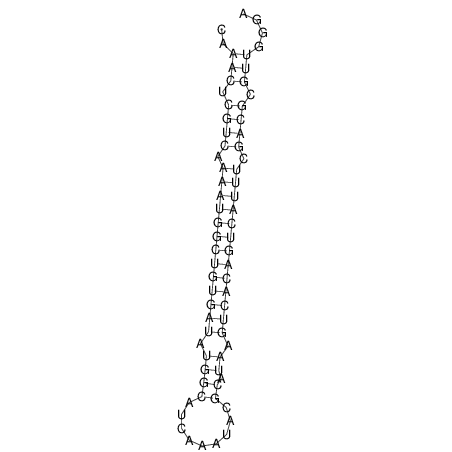
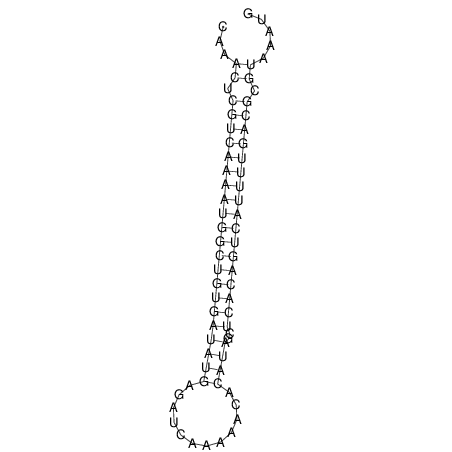
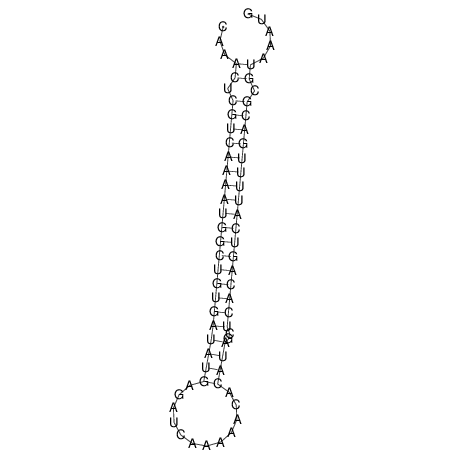
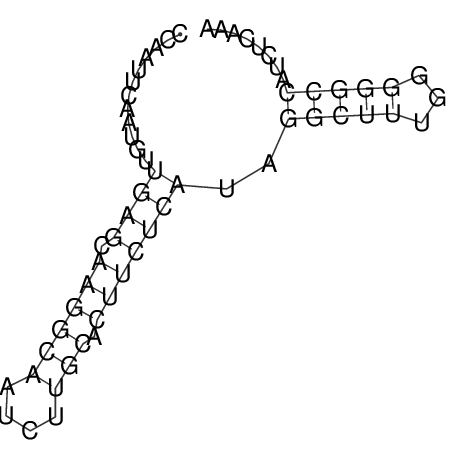
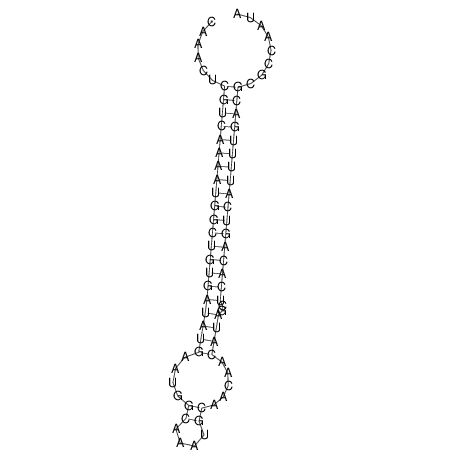
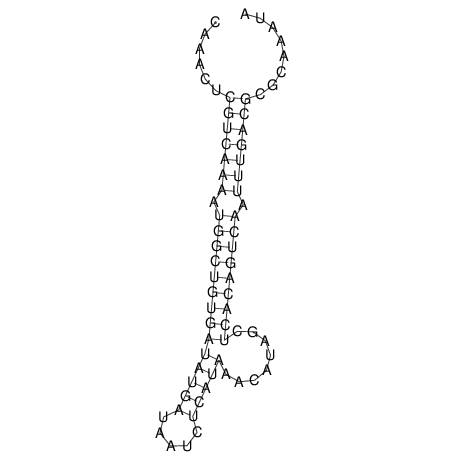
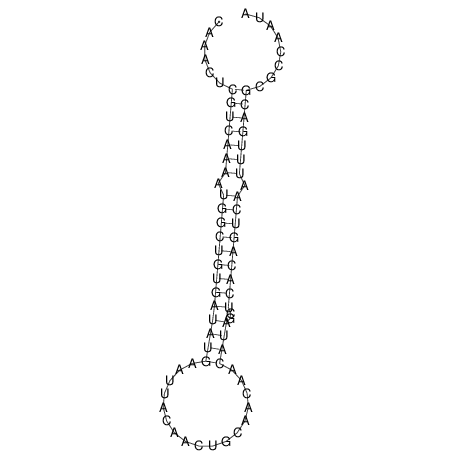
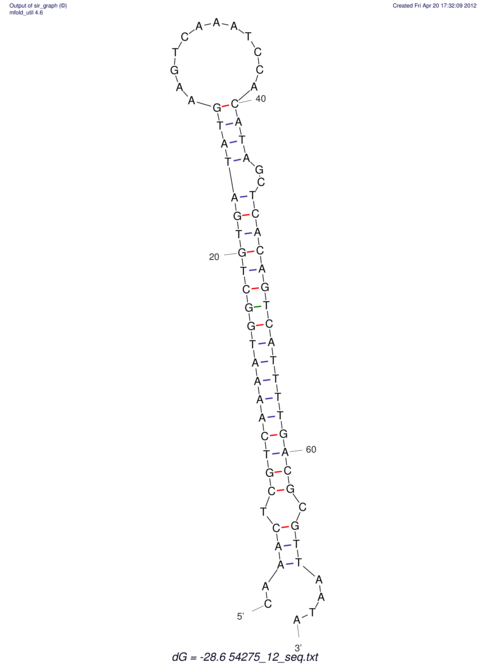
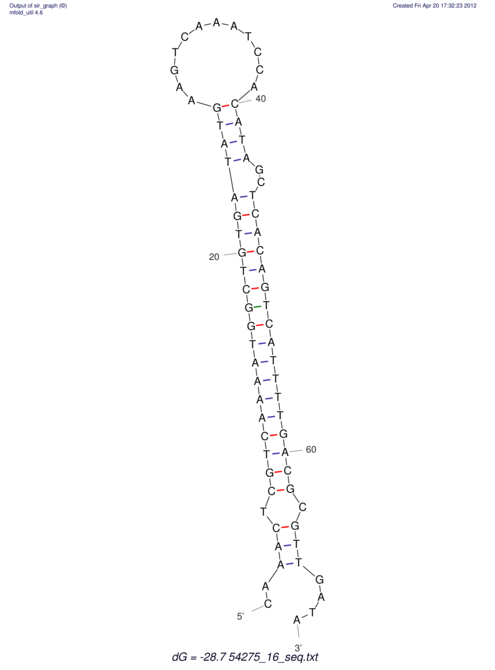
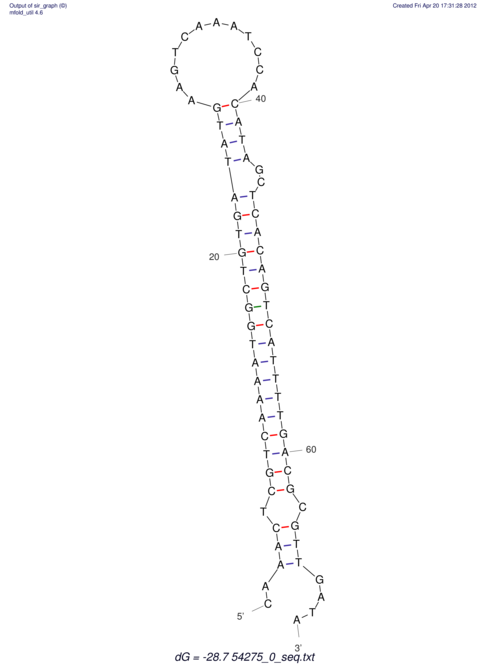
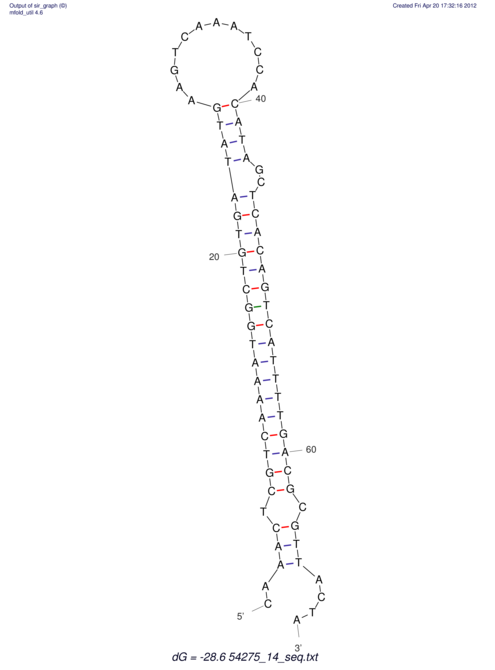
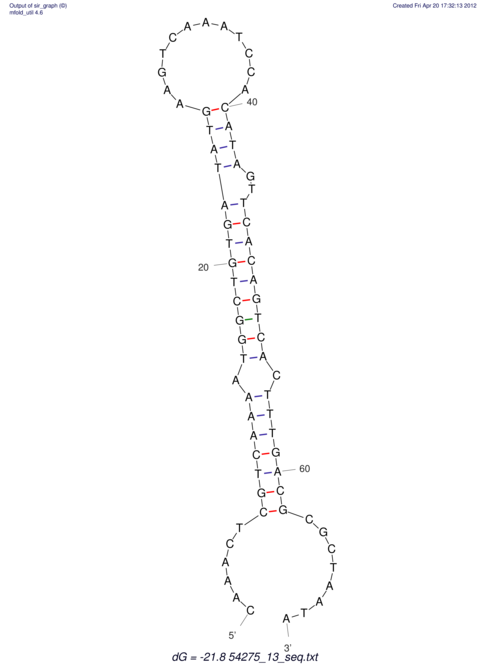
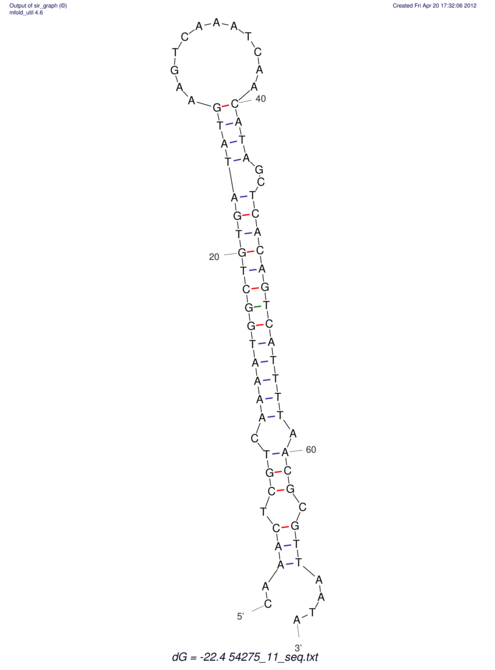
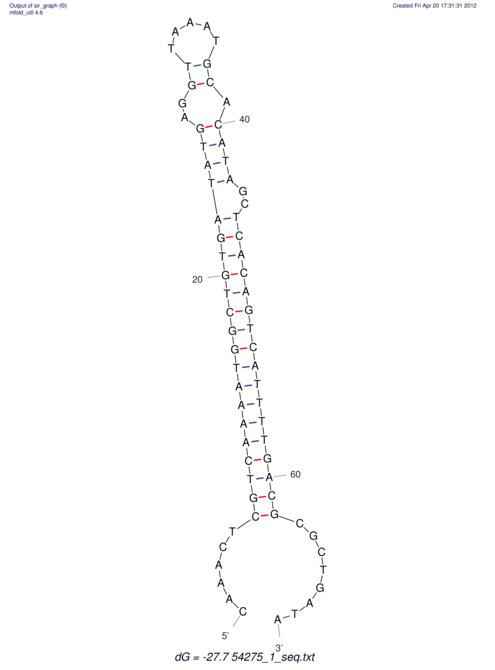
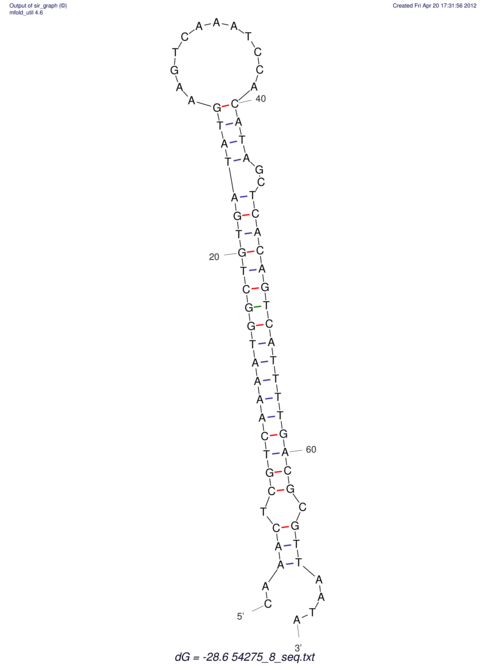
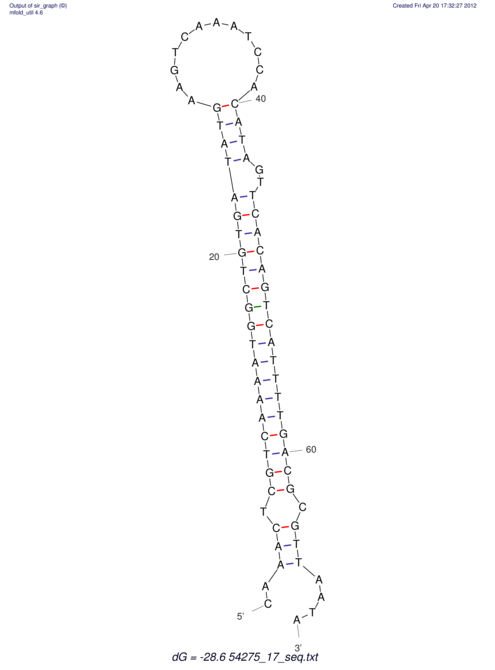
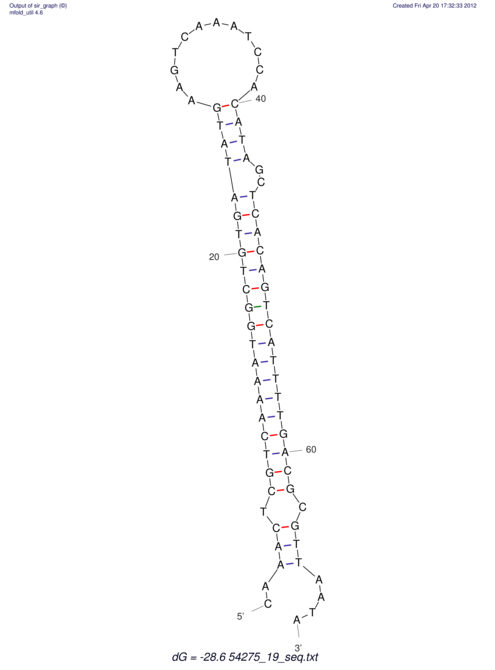
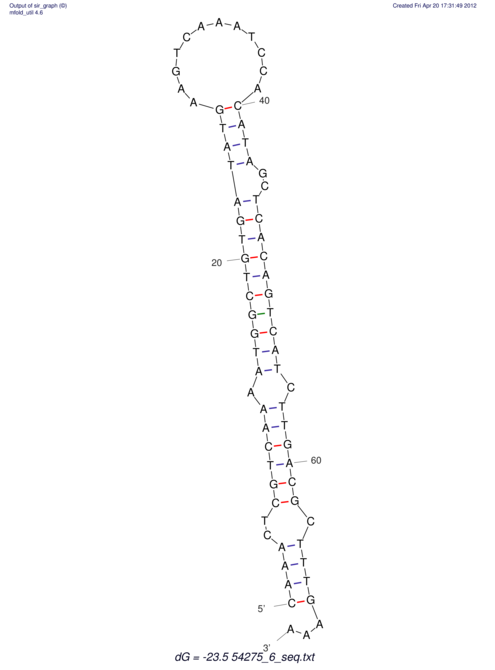
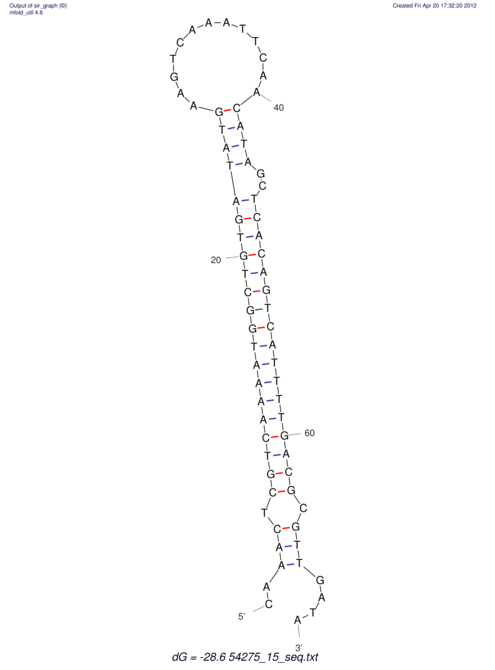
| A-----GACTGGC-CAGCAAACTCGTCAAAATGGCTGTGATATGAA-G-TCAA----ATCCACATAGCTCACAGTCATTTTGACGCGTT-ACTAGACAGCAG-G--GG-----------TAAG | Size | Hit Count | Total Norm | V046
Embryo | V052
Head | V120
Male body | V058
Head | M026
Head |
| ......................................................................ACAGCCATTTTGACGCGTT-................................. | 19 | 9 | 1.11 | 0.22 | 0.00 | 0.00 | 0.89 | 0.00 |
| ......................................................................ACAGCCATTTTGACGCGT................................... | 18 | 9 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ......................................................................ACAGCCATTTTGACGAGTT-A................................ | 20 | 9 | 0.78 | 0.56 | 0.11 | 0.00 | 0.11 | 0.00 |
| ..................................................................TCTCACAGCCATTTTGACGCGT................................... | 22 | 7 | 0.86 | 0.43 | 0.14 | 0.00 | 0.29 | 0.00 |
| ..................................................................TATCACAGTCATTTTGACGCGT................................... | 22 | 7 | 0.86 | 0.29 | 0.00 | 0.00 | 0.57 | 0.00 |
| ..................................................................TCTCACAGCCATTTTGACGCGTT-................................. | 23 | 7 | 0.86 | 0.14 | 0.00 | 0.00 | 0.71 | 0.00 |
| ..................................................................TATCACAGTCATTTTGACGCGTT-................................. | 23 | 7 | 0.57 | 0.29 | 0.00 | 0.00 | 0.29 | 0.00 |
| ......................................................................ACAGACATTTTGACGAGTT-................................. | 19 | 9 | 0.33 | 0.11 | 0.00 | 0.00 | 0.22 | 0.00 |
| ..................................................................TCTCACAGTCATTTTGACGAGT................................... | 22 | 7 | 0.29 | 0.00 | 0.00 | 0.00 | 0.29 | 0.00 |
| .......................................................................CAGCCATTTTGACGAGTT-A................................ | 19 | 9 | 0.22 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................GCTCACAGCCATTTTGACGAGT................................... | 22 | 7 | 0.29 | 0.29 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................ACAGACATTTTGACGAGT................................... | 18 | 9 | 0.22 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 |
| ....................................................................TCACAGCCATTTTGACGAGTT-A................................ | 22 | 9 | 0.22 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................ACAGCCATTTTGATGCGTT-................................. | 19 | 9 | 0.11 | 0.00 | 0.00 | 0.00 | 0.11 | 0.00 |
| ......................................................................ACAGGCATTTTGACGAGT................................... | 18 | 9 | 0.11 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................GCTCACAGCCATTTTGACGAGTT-................................. | 23 | 7 | 0.14 | 0.00 | 0.14 | 0.00 | 0.00 | 0.00 |
| ......................................................................ACAGCCATTTTGACGTGT................................... | 18 | 9 | 0.11 | 0.00 | 0.00 | 0.00 | 0.11 | 0.00 |
| ..................................................................AATCACAGTCATTTTGACGCGT................................... | 22 | 2 | 0.50 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 |
| ......................................................................ACAGACATTTTGACGCGT................................... | 18 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ..................................................................GATCACAGCCATTTTGACGCGT................................... | 22 | 7 | 0.14 | 0.14 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................GATCACAGCCATTTTGACGCGTT-................................. | 23 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ..................................................................GATCACAGTCATTTTGACGTGT................................... | 22 | 2 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 |
| ....................................................................TCACAGCCCTTTTGACGCGT................................... | 20 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| .....................................................................CACAGCCATTTTGACGCGT................................... | 19 | 9 | 0.11 | 0.00 | 0.00 | 0.00 | 0.11 | 0.00 |
| ......................................................................ACAGCCATTTTGACGCAT................................... | 18 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ......................................................................ACAGCCATTTTGCCGCGT................................... | 18 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ......................................................................ACAGTCATTTTGATGAGTT-................................. | 19 | 9 | 0.11 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................ACAGCCATTTTGACGGGTT-................................. | 19 | 9 | 0.11 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................ACAGTCATTTTGACGCGC................................... | 18 | 2 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................ACAGCCATTTTGACGCCT................................... | 18 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 | 0.00 |
| ......................................................................ACAGCCATTTTGACGGG.................................... | 17 | 9 | 0.11 | 0.00 | 0.00 | 0.00 | 0.11 | 0.00 |
| ...................................................................ATCACAGCCATTTTGACGCGTT-................................. | 22 | 9 | 0.11 | 0.00 | 0.00 | 0.00 | 0.11 | 0.00 |
| ..................................................................TCTCACAGTAATTTTGACGCGT................................... | 22 | 2 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................ACAGTCATTTTGACGAGTT-................................. | 19 | 9 | 0.11 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| ........................................................................AGCCATTTTGACGAGTT-A................................ | 18 | 9 | 0.11 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................TATCACAGTCATTTTGACG...................................... | 19 | 9 | 0.11 | 0.00 | 0.11 | 0.00 | 0.00 | 0.00 |
| ...........................................................................................CTAGACAGCGG-G--GG-----------AA.. | 16 | 2 | 0.50 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 |