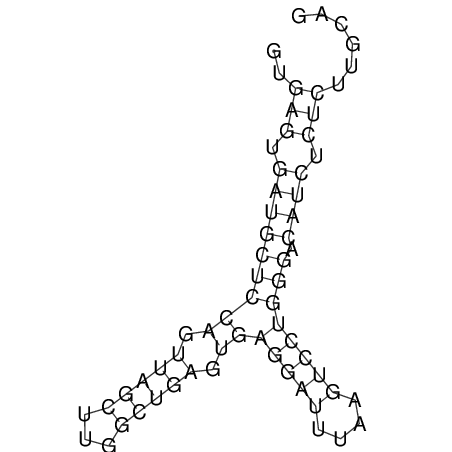
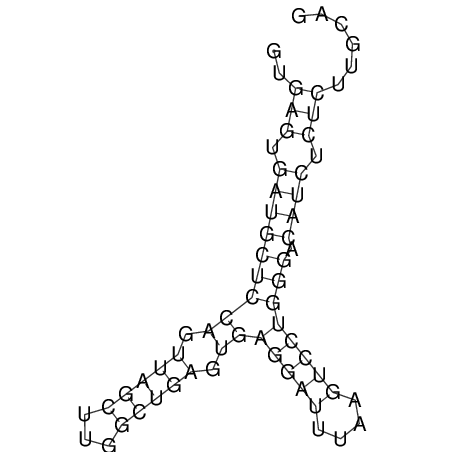

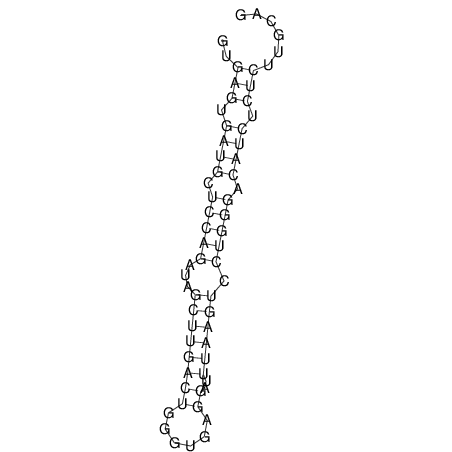
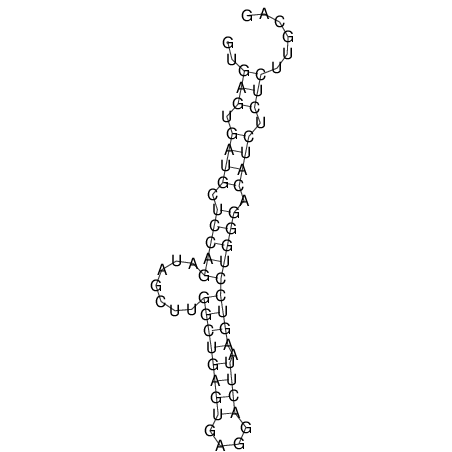
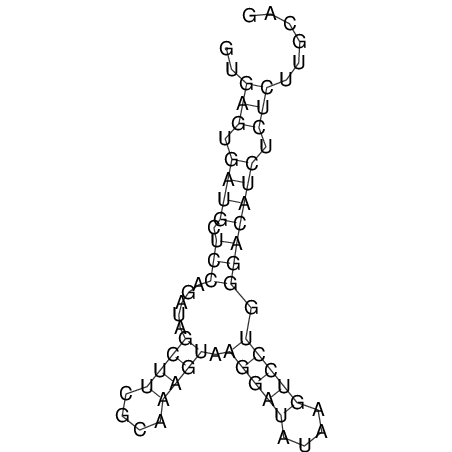
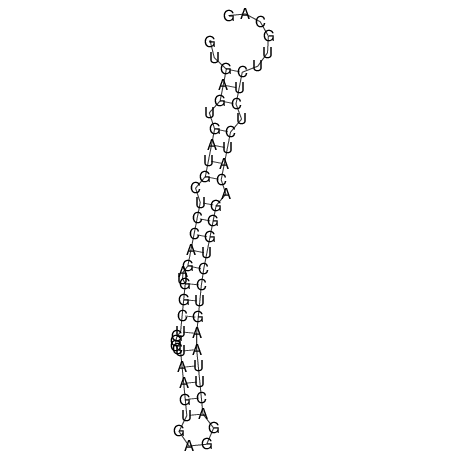
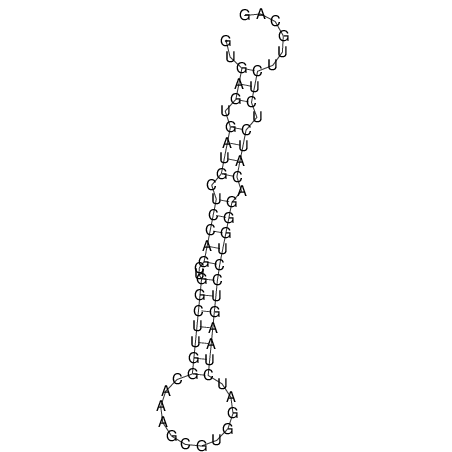
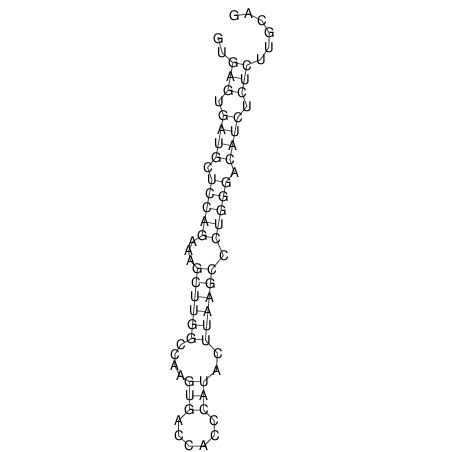

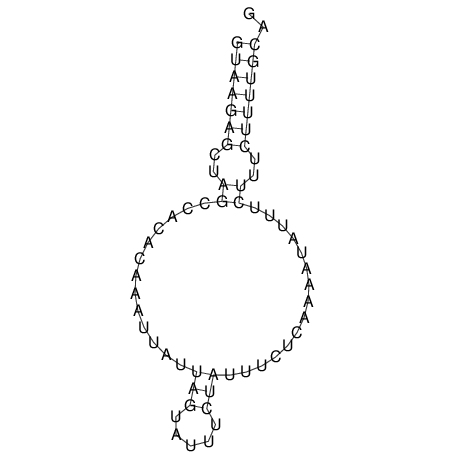

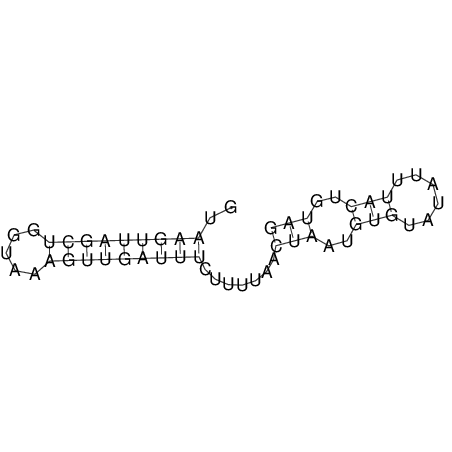
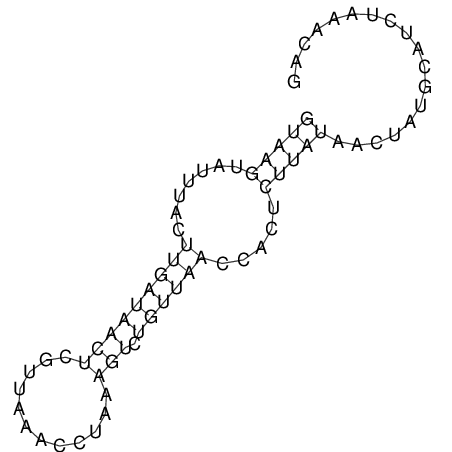
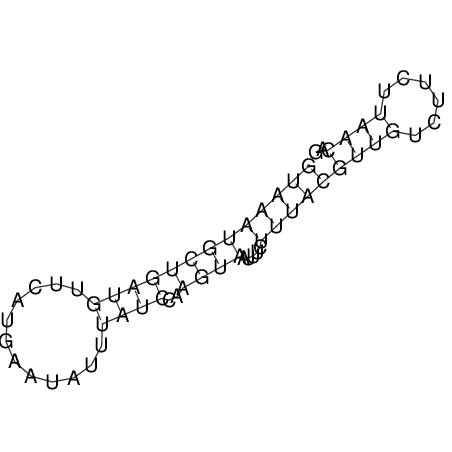

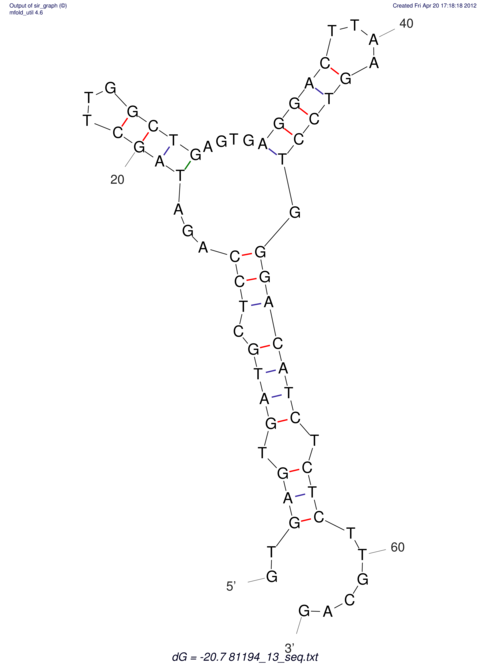

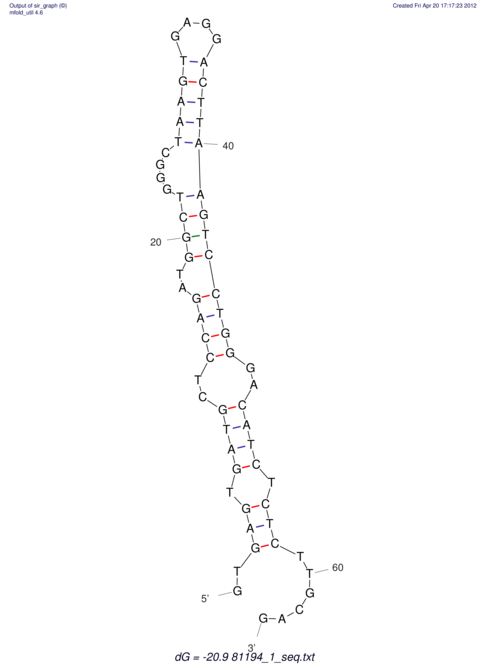
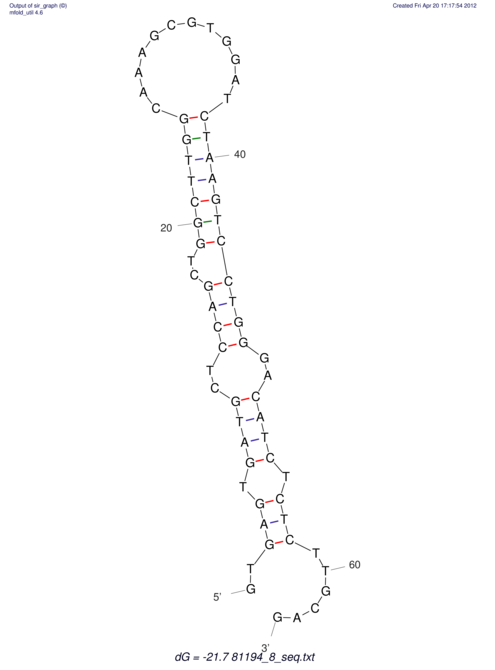
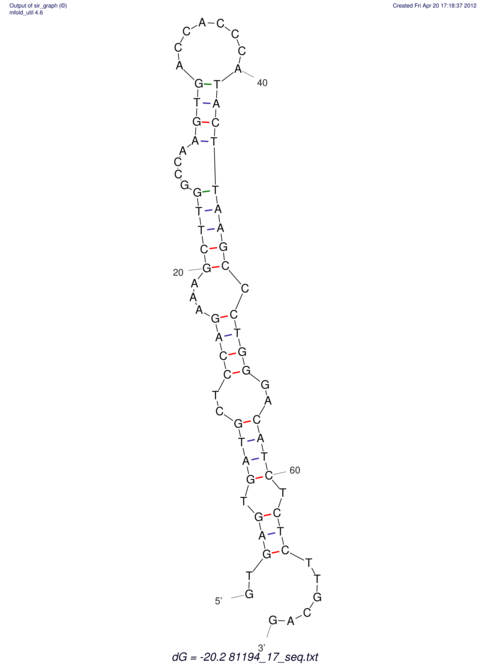


View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
dme-mir-1015 |
chr3R:20164954-20165017 + |
Known_mirbase |
|---|---|---|
| [View on miRBase] | [View on UCSC Genome Browser {Cornell Mirror}] | mirtron |
Legend:
mature strand at position 20164996-20165017Notes: The miRNA appears lost in dp4 - droGri2. The orthologs for these species look confident however they've accumulated too many substitutions to fold nicely.
| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr3R:20164939-20165032 + | 1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGTTAGCTTGGCTGAGTGAG-G--------A----TT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAAG | |
| droSim1 | chr3R:19998542-19998635 + | LASTZ | 1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGTTAGCTTGGCTGAGTGAG-G--------A----TT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTAAAAG |
| droSec1 | super_0:20503496-20503589 + | LASTZ | 1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGTTAGCTTGTCTGAGTGAG-A--------C----TT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTAAAAG |
| droYak2 | chr3R:26340571-26340664 - | LASTZ | 1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGATAGCTTGACTGGGTGAG-G--------A----TT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAAC |
| droEre2 | scaffold_4820:7895693-7895786 - | LASTZ | 1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGATAGCTTGGCTGAGTGAG-G--------A----CT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAAATCCTAAAAC |
| droEug1 | scf7180000409798:215829-215922 - | LASTZ | 5 | TAATGGGCTACAATGGTGAGTG----ATGCTCCAGATAGCTTCGCAAAGTAAG-G--------A----TA------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTAAGAC |
| droBia1 | scf7180000302136:526695-526788 + | LASTZ | 4 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGATGGCTGGGCTAAGTGAG-G--------A----CT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAGC |
| droTak1 | scf7180000415769:2262017-2262110 - | LASTZ | 4 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGCTGGCTTGGCAAAGCGTG-G--------A----TC------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTCAAGC |
| droEle1 | scf7180000491008:477705-477804 + | LASTZ | 4 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGAAAGCTTGGCCAAGTGAC-C--------A----CCCATACTTAAGCCCTGGGAC-----ATCTCTCTTGCAGTGAAGGTCCTGAAGC |
| droRho1 | scf7180000779477:268222-268317 + | LASTZ | 4 | TCATGGGATACGACGGTGAGTG----ATGCTCCAGATAGCTTGGCTAAGTGAG-G--------A----TTAA----TAAGCCCTGGGAC-----ATCTCTCTTGCAGTGAAGGTCCTGAAGC |
| droFic1 | scf7180000453912:1339925-1340020 + | LASTZ | 5 | TCATGGGCTACCAAGGTGAGTG----ATGCTCCAGATAACTTGGAAAAGTGGG-A--------ATACGGA------TAAGTCC--GGAC-----TTCTCTCTTGCAGTCAAGATTCTGAAAT |
| droKik1 | scf7180000302387:55058-55154 + | LASTZ | 4 | TGATGGGCTATGACGGTGAGTG----ATGCTCCAGATAGCTTAGTTTAAAAAAAG--------A----TTCA----AAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAGC |
| droAna3 | scaffold_13088:204245-204339 + | LASTZ | 1 | TCATGGGCTACGACAGTGAGTG----ATGCTCCAGATAGGTTGGCTTAGGAAGTC--------A----TT------AAAGCCCTGGGAT-----GTCTCTCTTGCAGTTAAGATCCTGAAAC |
| droBip1 | scf7180000396413:258464-258558 + | LASTZ | 4 | TCATGGGCTACGATAGTGAGTG----ATGCTCTAGATAGGTTAGCTGATGAAG-T--------C---TTT------CAAGCCCTAGGTC-----ATCTCTCTTGCAGTTAAAATCCTGAAAC |
| dp4 | chr2:18646206-18646298 - | LASTZ | 5 | TGATGGGCTACGATGGTAAGAG----CTAGCCA-CACAAATTA------TTAG-T--------A----TTTC----TATTTCTCAAAATATTT-CTTTCTTTTGCAGTCAAGATACTAAAGC |
| droPer1 | super_3:1376406-1376498 - | LASTZ | 5 | TGATGGGCTACGATGGTAAGAG----CTAGCCA-CACAAATTA------TTAG-T--------A----TTTC----TATTTCTCAAAATATTC-CTTTCTTTTGCAGTCAAGATCCTAAAGC |
| droWil1 | scaffold_181108:4541725-4541808 - | LASTZ | 5 | TAATGGGTTATGACAGTAAGTTA--GCTGGTAAAGTTGATTTCT------------------------TT------TAACTAATGT-GT-----ATATTTACTGTAGTAAGAATTTTAAAGC |
| droVir3 | scaffold_13047:3497490-3497590 + | LASTZ | 5 | TTGTGGGCTACGAAAGTAAGTA----T-TTACTTGATAACTCGTT------AA-ACCTAAAGTC----TGTT----AACCACTCTTATAACTATG-CATCTAAACAGTTAAGGTACTAAAGA |
| droMoj3 | scaffold_6540:20508341-20508431 + | LASTZ | 5 | TAATGGGCTACGATGGTAAATG-CTGATGTTCATGAATATTTATCCAAGTAAT-T--------T----------------CTTTACGTT-----GTCTTCTTAACAGTTAAGGTTTTGAAAA |
| droGri2 | scaffold_14906:156637-156731 + | LASTZ | 5 | TGGTGGGCTATGATAGTGAGTTG---ATTCTATTGT--AATAATC-AAATC-G-T--------T----TTTG----TTATTTTATAAAC---ATGCCATTTTAACAGTCAAGATACTCAAGA |
| Species | Alignment |
| dm3 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGTTAGCTTGGCTGAGTGAG-G--------A----TT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAAG |
| droSim1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGTTAGCTTGGCTGAGTGAG-G--------A----TT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTAAAAG |
| droSec1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGTTAGCTTGTCTGAGTGAG-A--------C----TT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTAAAAG |
| droYak2 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGATAGCTTGACTGGGTGAG-G--------A----TT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAAC |
| droEre2 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGATAGCTTGGCTGAGTGAG-G--------A----CT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAAATCCTAAAAC |
| droEug1 | TAATGGGCTACAATGGTGAGTG----ATGCTCCAGATAGCTTCGCAAAGTAAG-G--------A----TA------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTAAGAC |
| droBia1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGATGGCTGGGCTAAGTGAG-G--------A----CT------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAGC |
| droTak1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGCTGGCTTGGCAAAGCGTG-G--------A----TC------TAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTCAAGC |
| droEle1 | TCATGGGCTACGACGGTGAGTG----ATGCTCCAGAAAGCTTGGCCAAGTGAC-C--------A----CCCATACTTAAGCCCTGGGAC-----ATCTCTCTTGCAGTGAAGGTCCTGAAGC |
| droRho1 | TCATGGGATACGACGGTGAGTG----ATGCTCCAGATAGCTTGGCTAAGTGAG-G--------A----TTAA----TAAGCCCTGGGAC-----ATCTCTCTTGCAGTGAAGGTCCTGAAGC |
| droFic1 | TCATGGGCTACCAAGGTGAGTG----ATGCTCCAGATAACTTGGAAAAGTGGG-A--------ATACGGA------TAAGTCC--GGAC-----TTCTCTCTTGCAGTCAAGATTCTGAAAT |
| droKik1 | TGATGGGCTATGACGGTGAGTG----ATGCTCCAGATAGCTTAGTTTAAAAAAAG--------A----TTCA----AAAGTCCTGGGAC-----ATCTCTCTTGCAGTCAAGATCCTGAAGC |
| droAna3 | TCATGGGCTACGACAGTGAGTG----ATGCTCCAGATAGGTTGGCTTAGGAAGTC--------A----TT------AAAGCCCTGGGAT-----GTCTCTCTTGCAGTTAAGATCCTGAAAC |
| droBip1 | TCATGGGCTACGATAGTGAGTG----ATGCTCTAGATAGGTTAGCTGATGAAG-T--------C---TTT------CAAGCCCTAGGTC-----ATCTCTCTTGCAGTTAAAATCCTGAAAC |
| dp4 | TGATGGGCTACGATGGTAAGAG----CTAGCCA-CACAAATTA------TTAG-T--------A----TTTC----TATTTCTCAAAATATTT-CTTTCTTTTGCAGTCAAGATACTAAAGC |
| droPer1 | TGATGGGCTACGATGGTAAGAG----CTAGCCA-CACAAATTA------TTAG-T--------A----TTTC----TATTTCTCAAAATATTC-CTTTCTTTTGCAGTCAAGATCCTAAAGC |
| droWil1 | TAATGGGTTATGACAGTAAGTTA--GCTGGTAAAGTTGATTTCT------------------------TT------TAACTAATGT-GT-----ATATTTACTGTAGTAAGAATTTTAAAGC |
| droVir3 | TTGTGGGCTACGAAAGTAAGTA----T-TTACTTGATAACTCGTT------AA-ACCTAAAGTC----TGTT----AACCACTCTTATAACTATG-CATCTAAACAGTTAAGGTACTAAAGA |
| droMoj3 | TAATGGGCTACGATGGTAAATG-CTGATGTTCATGAATATTTATCCAAGTAAT-T--------T----------------CTTTACGTT-----GTCTTCTTAACAGTTAAGGTTTTGAAAA |
| droGri2 | TGGTGGGCTATGATAGTGAGTTG---ATTCTATTGT--AATAATC-AAATC-G-T--------T----TTTG----TTATTTTATAAAC---ATGCCATTTTAACAGTCAAGATACTCAAGA |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
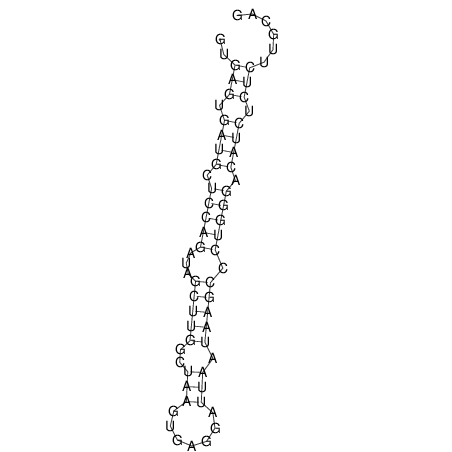
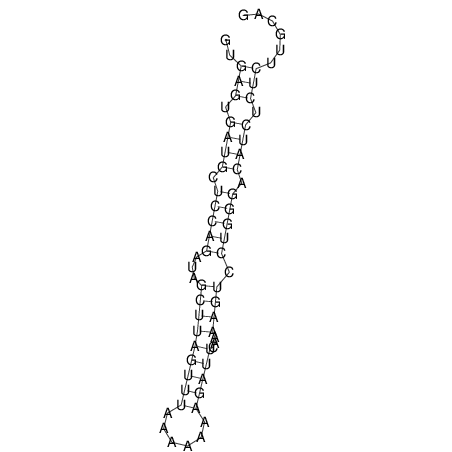
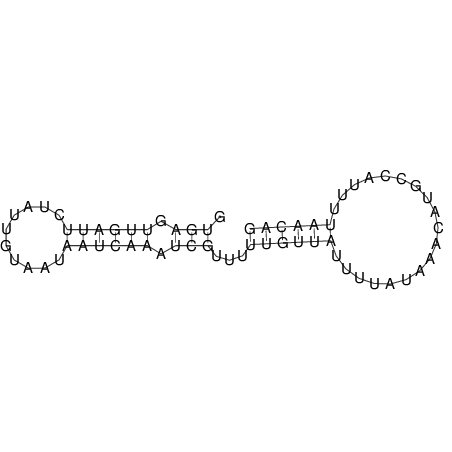
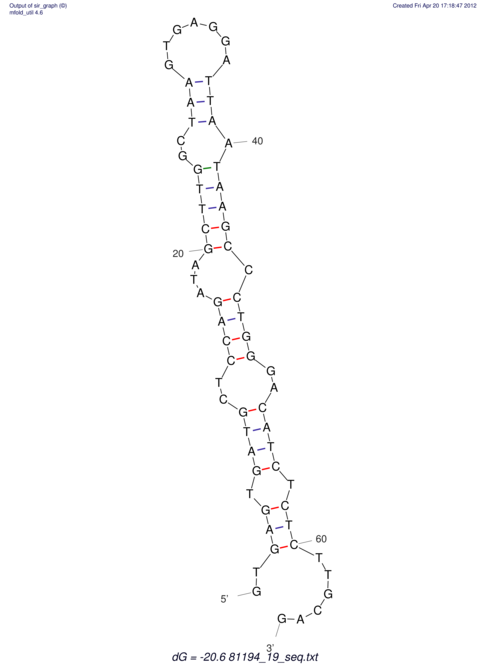
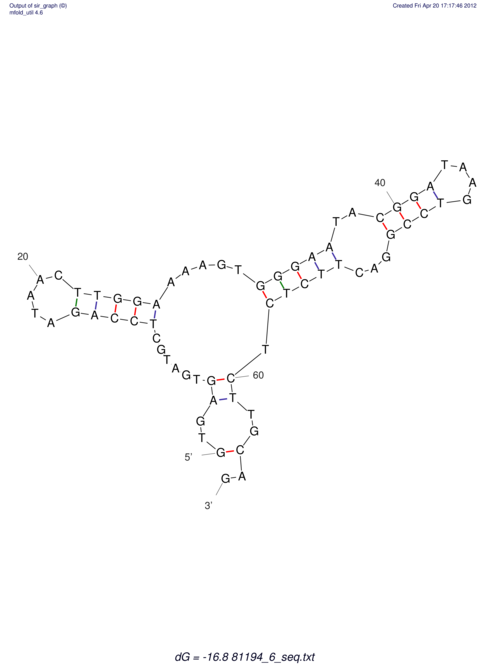
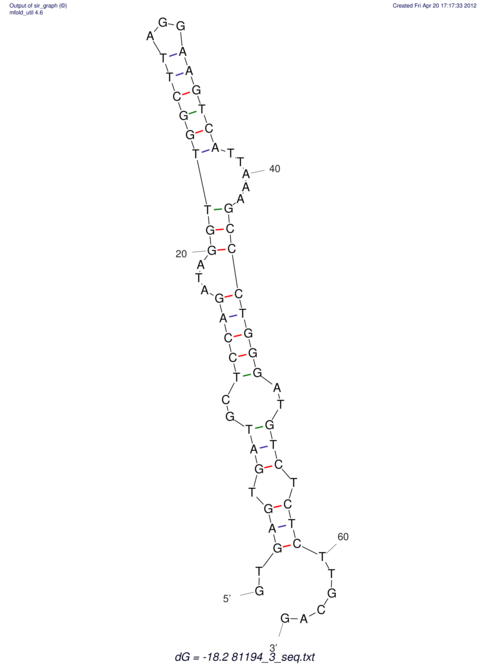
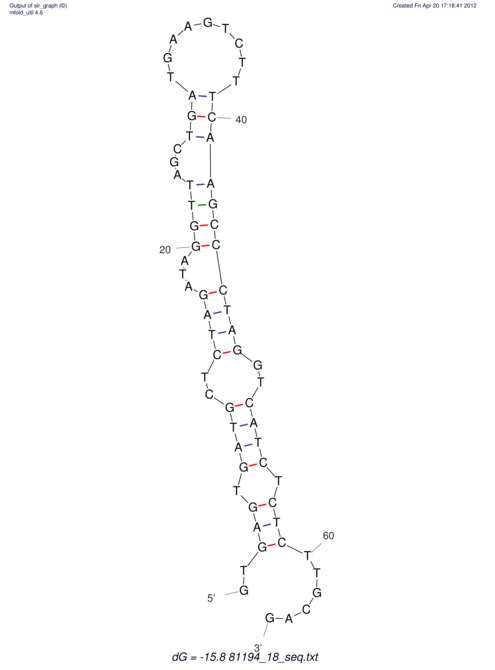
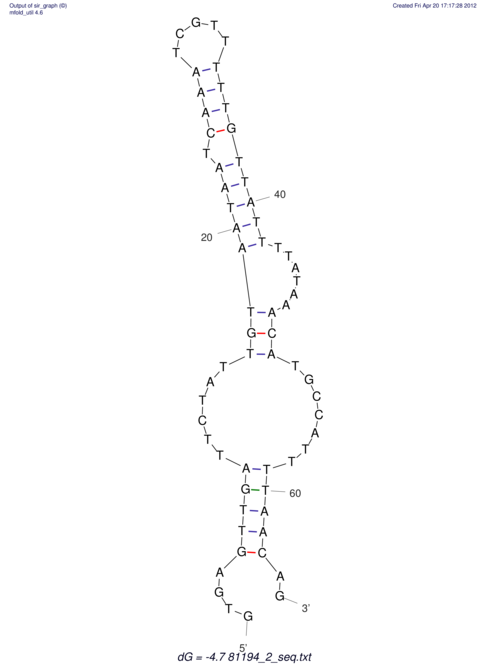
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
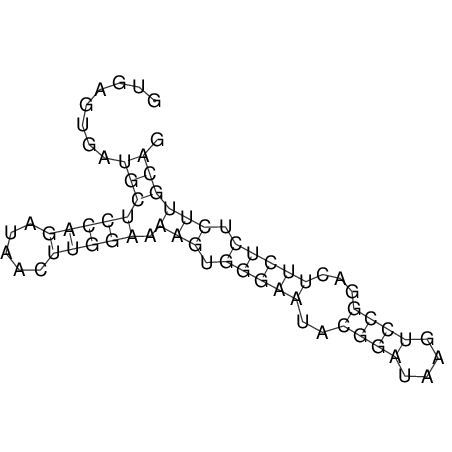
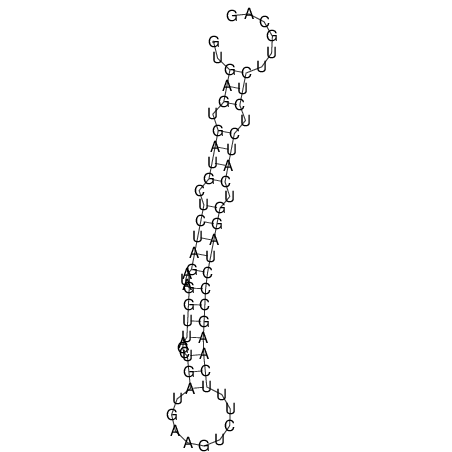
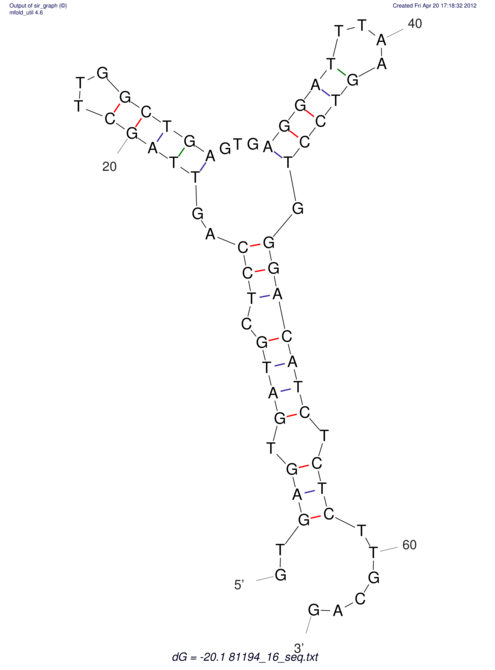
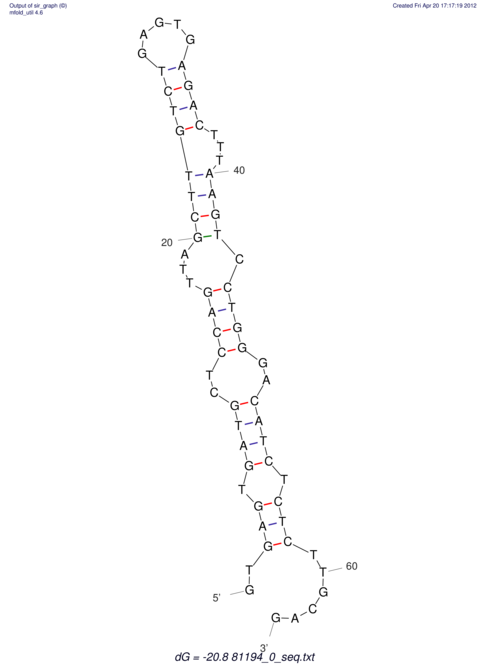
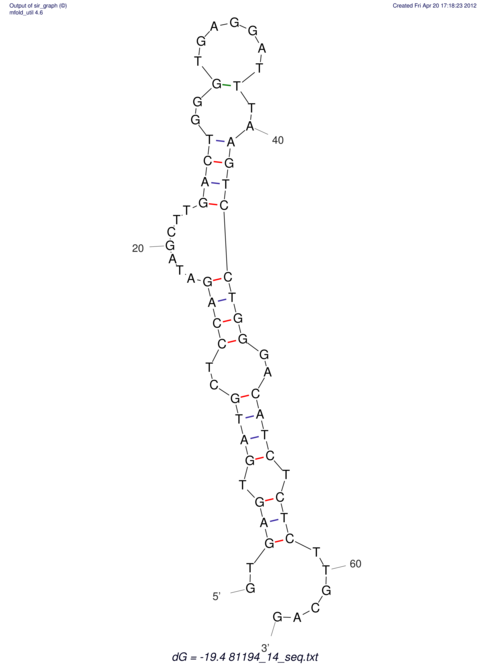
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:18:52 EDT