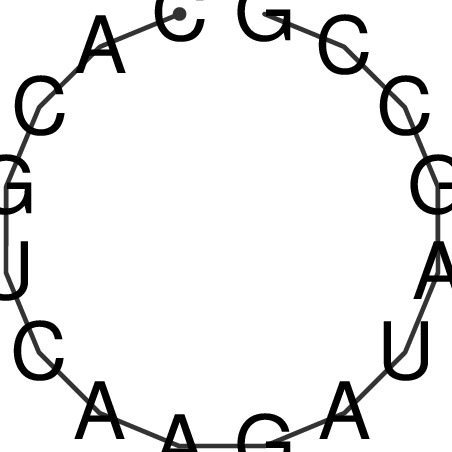
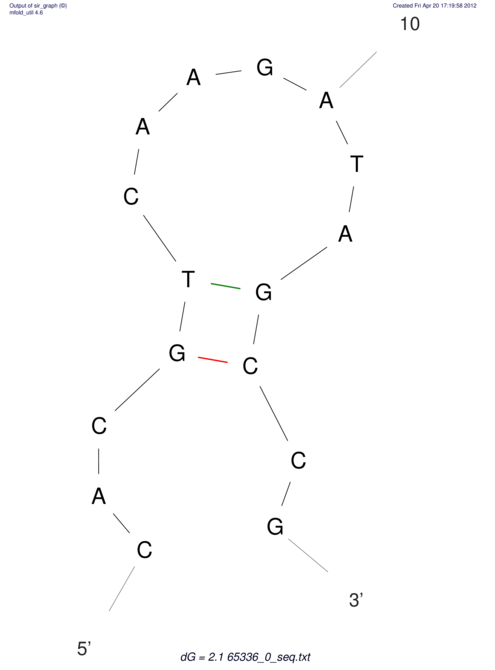
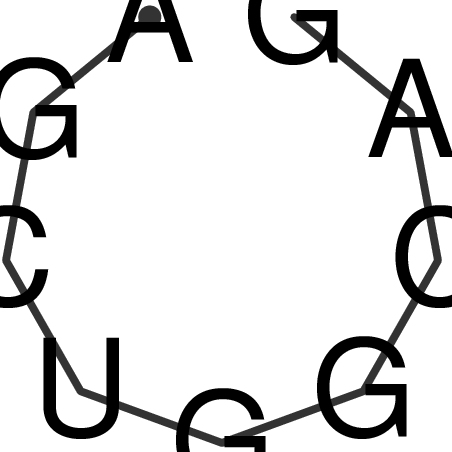
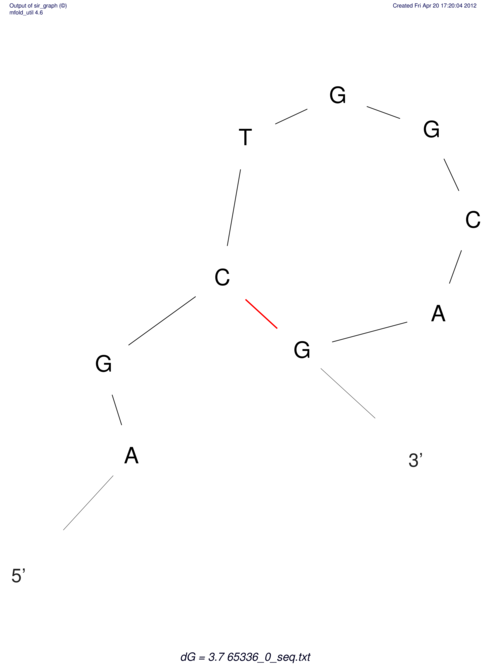
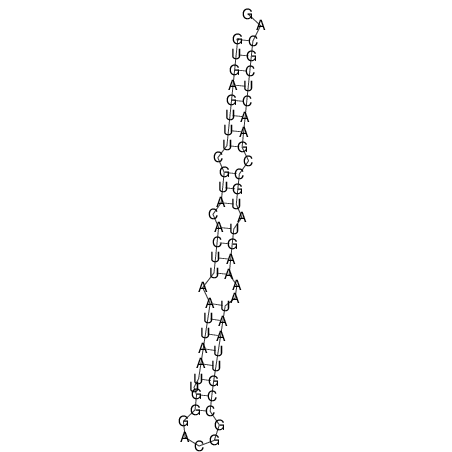
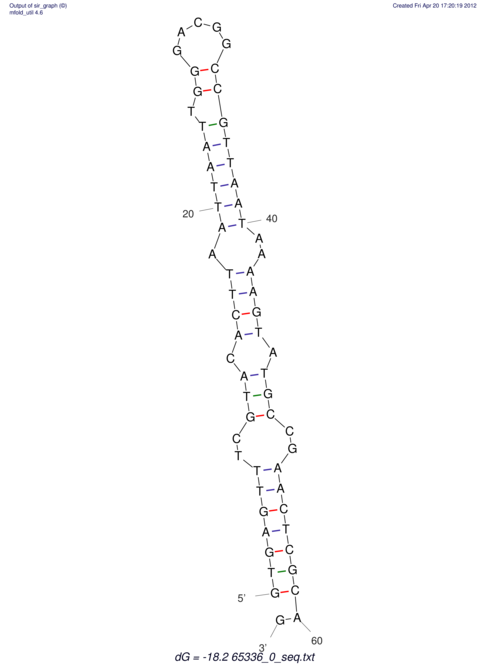
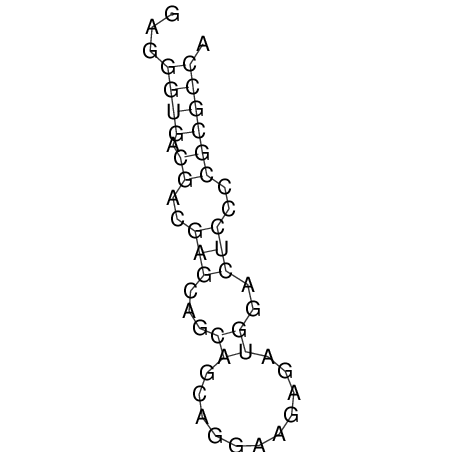
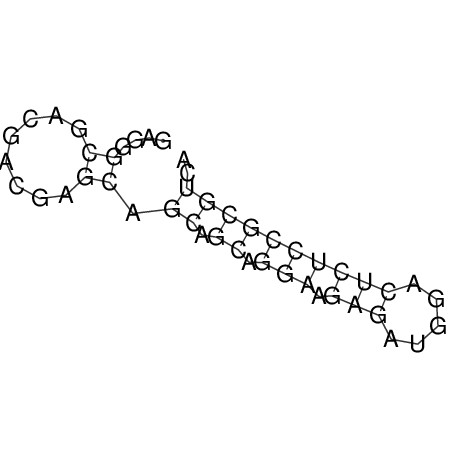
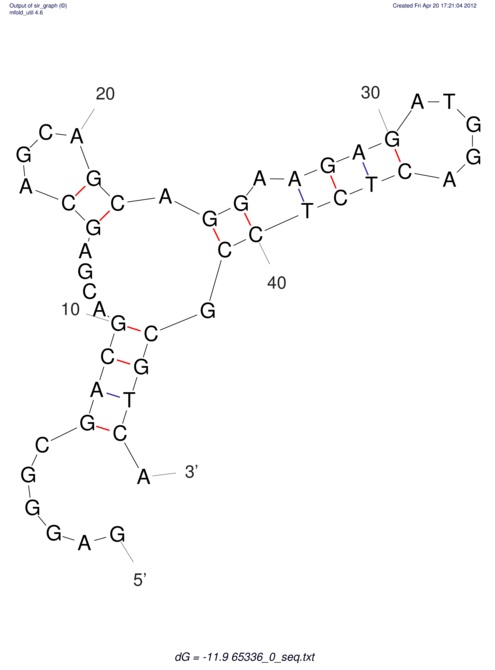
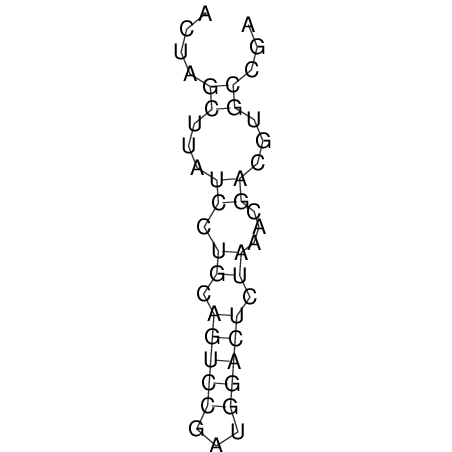
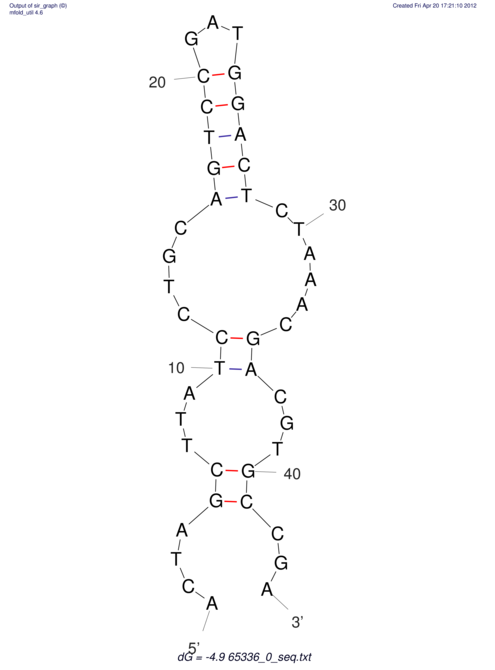
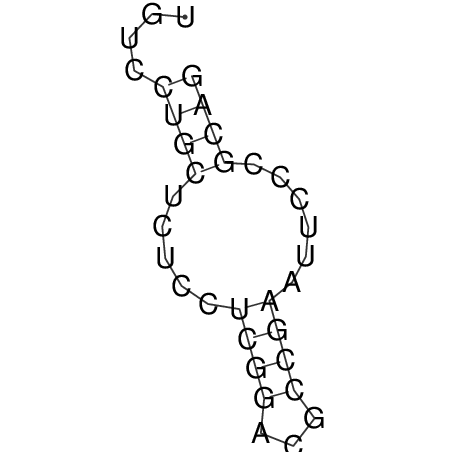
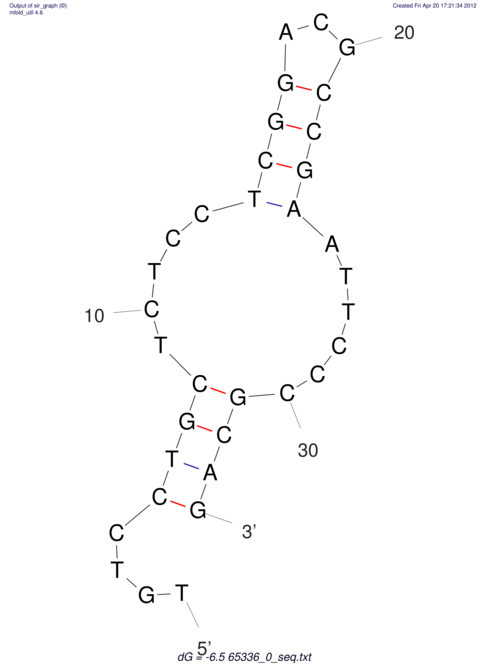
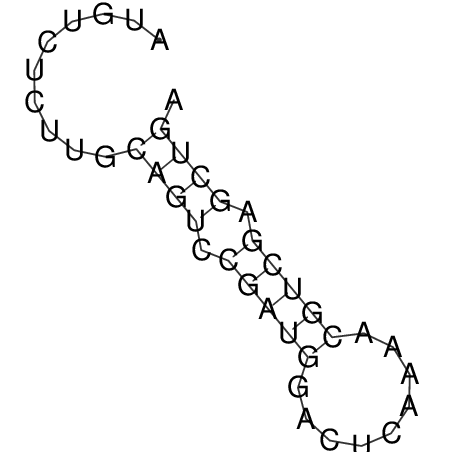
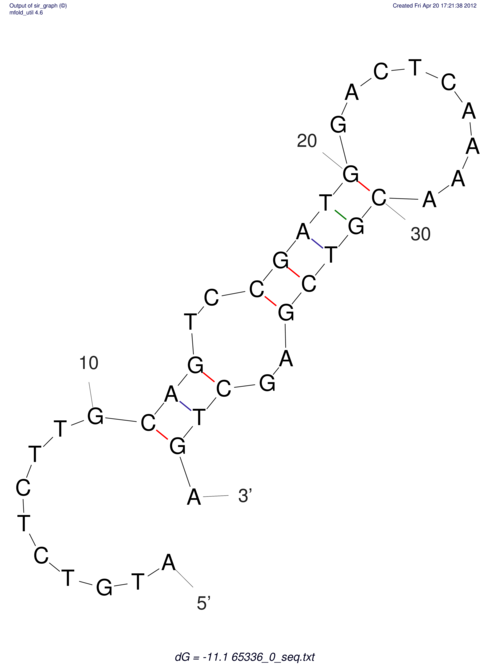
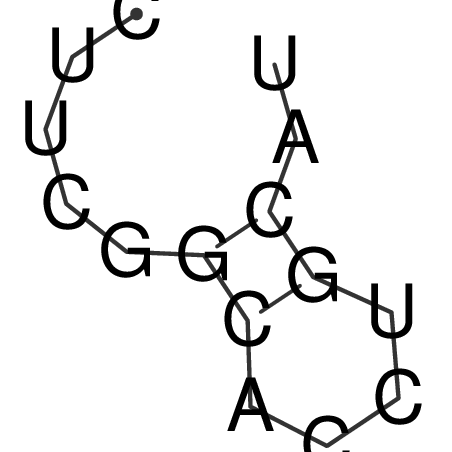
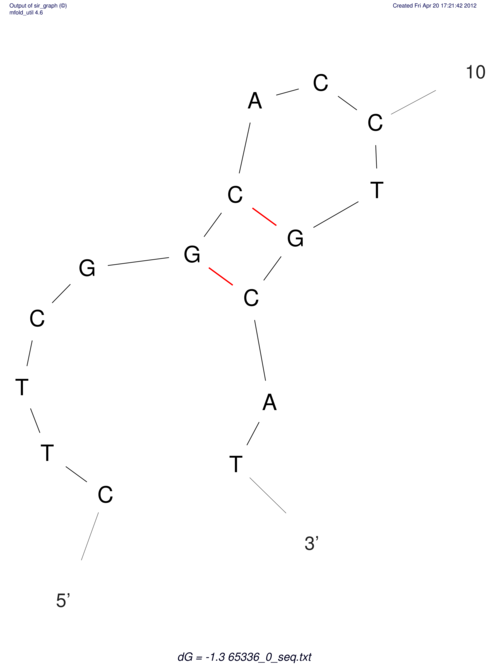
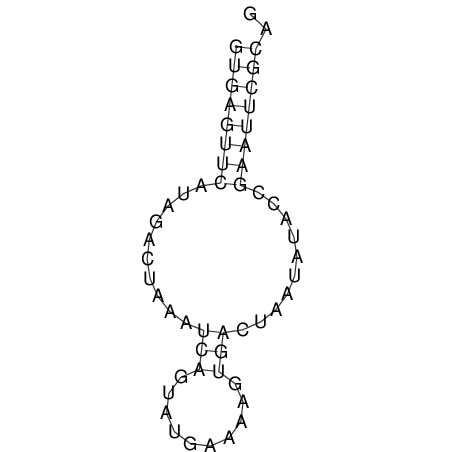
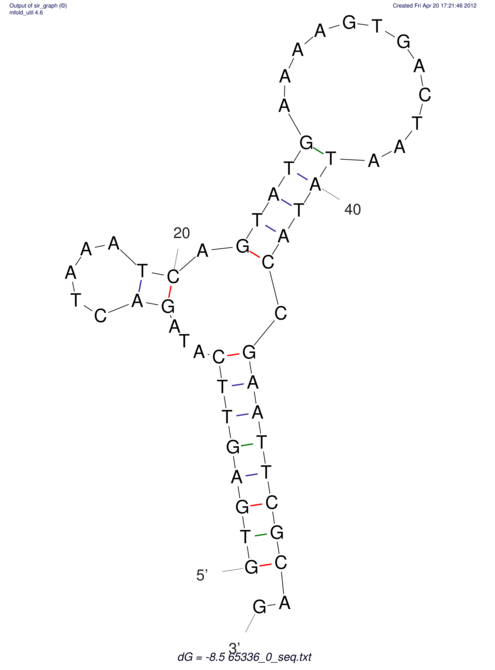
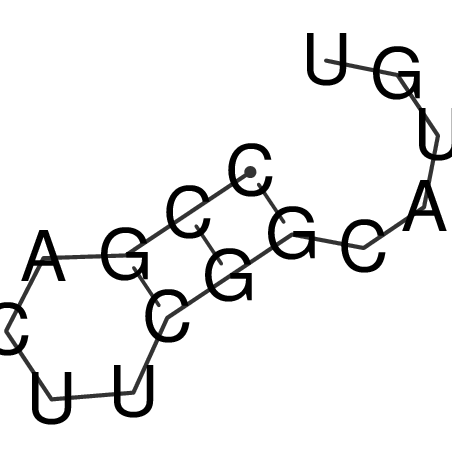
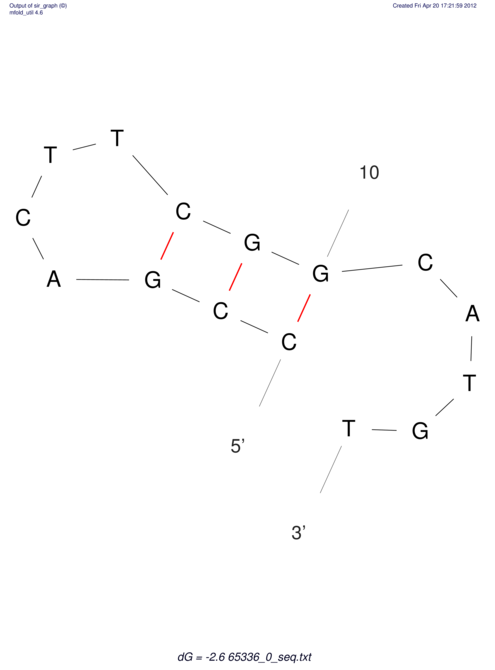
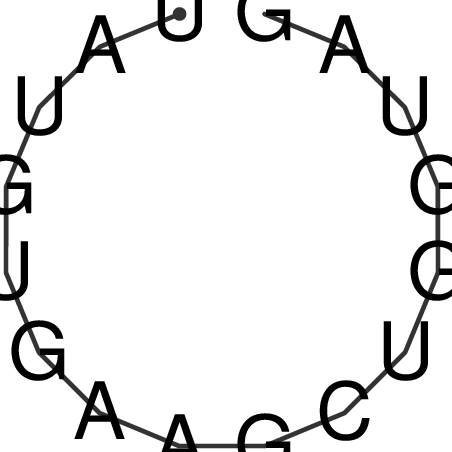
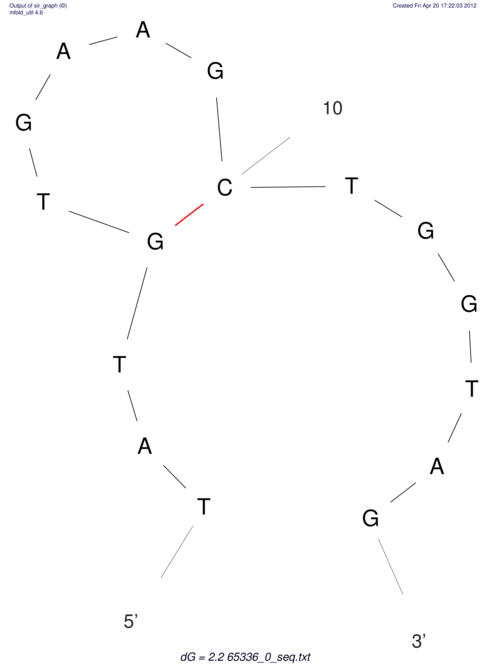
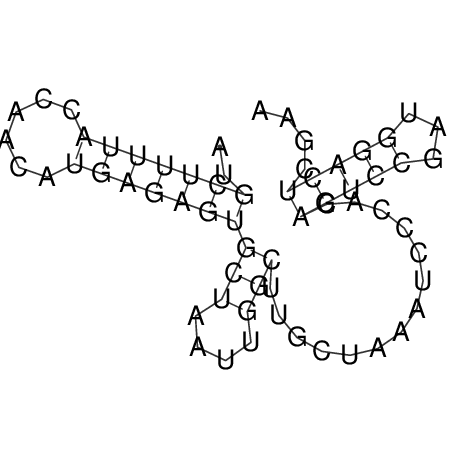
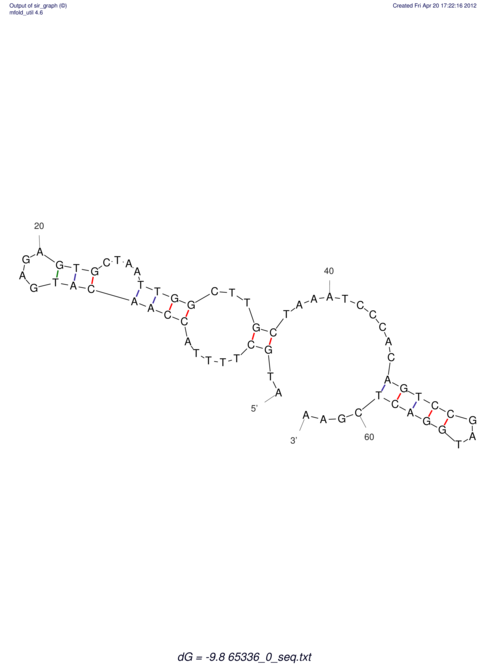
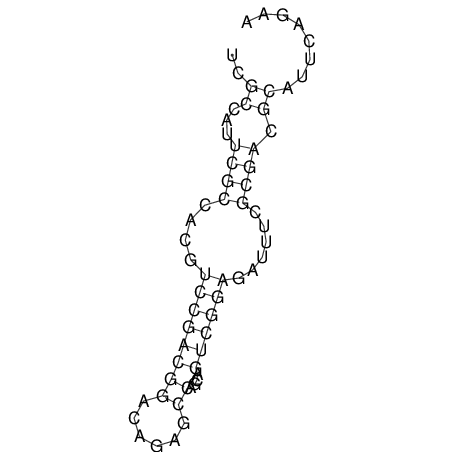
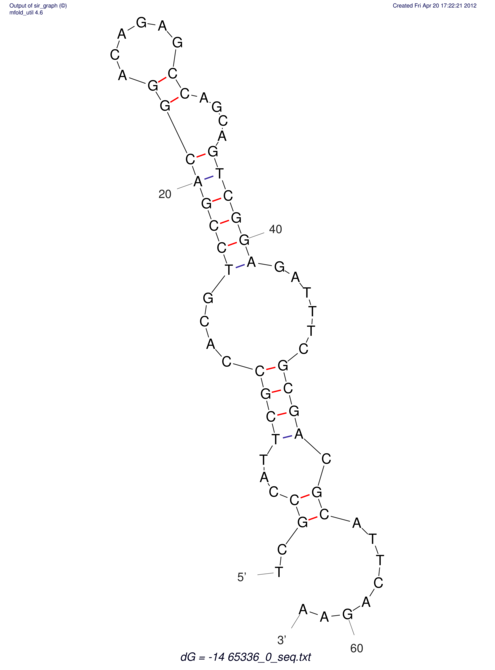
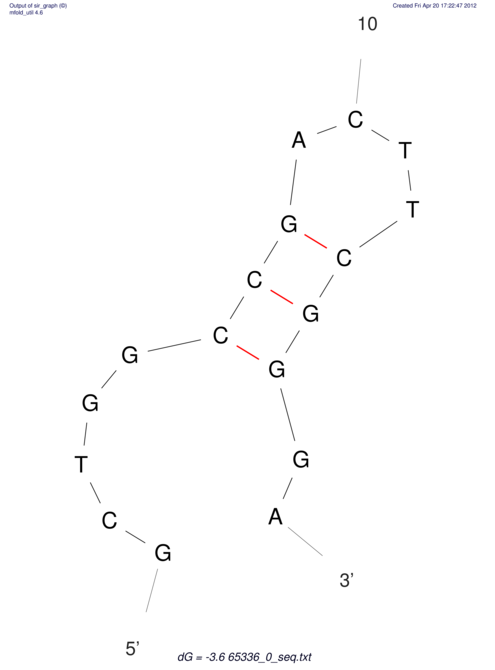
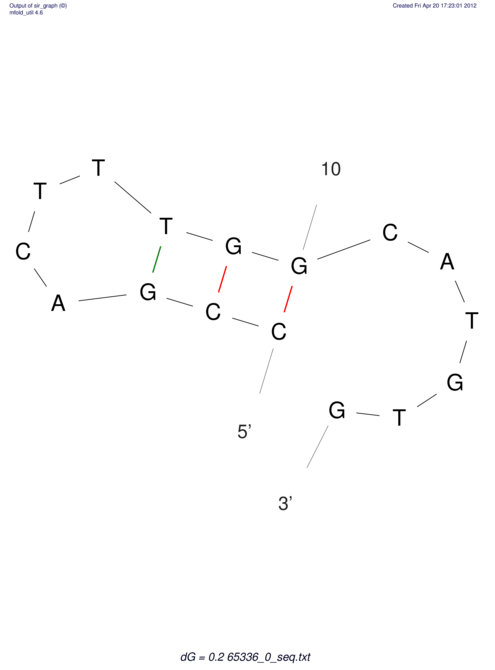
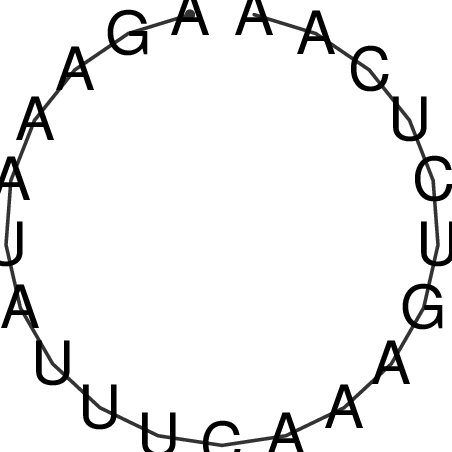
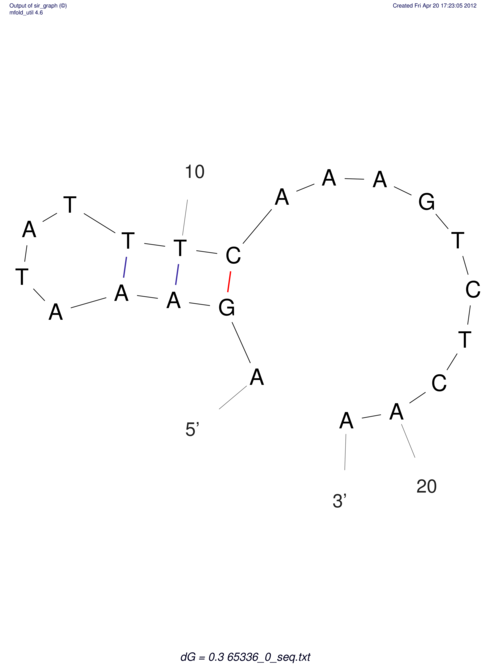
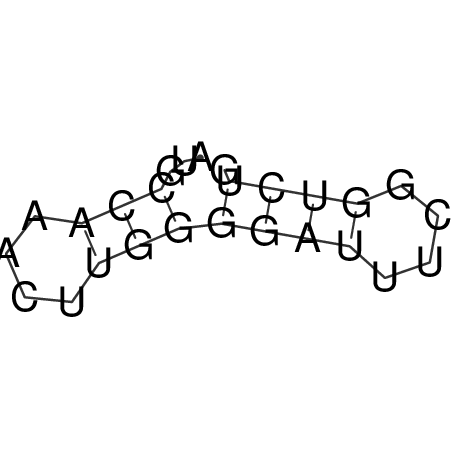
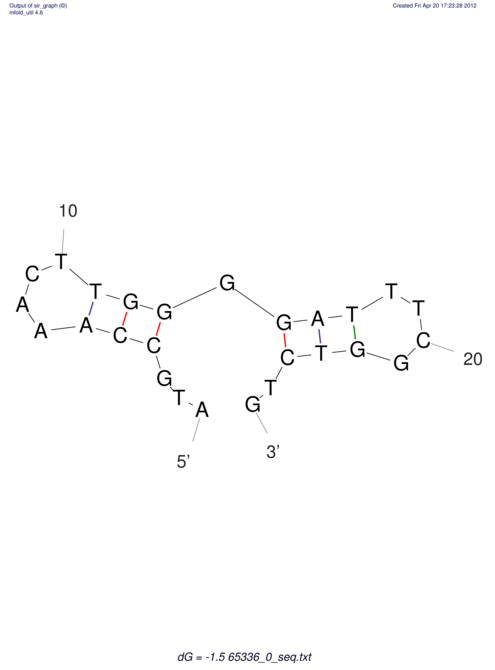
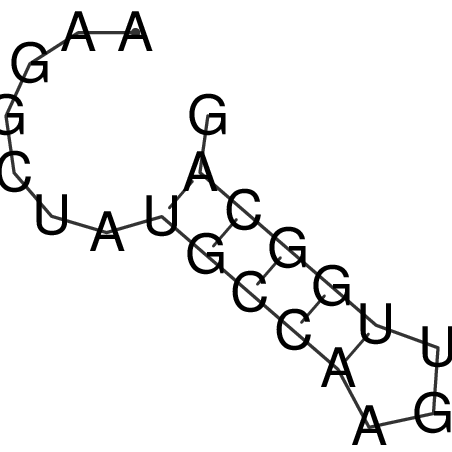
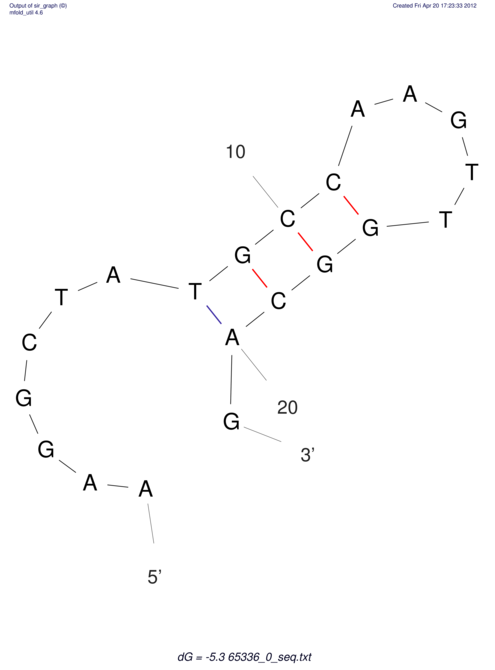
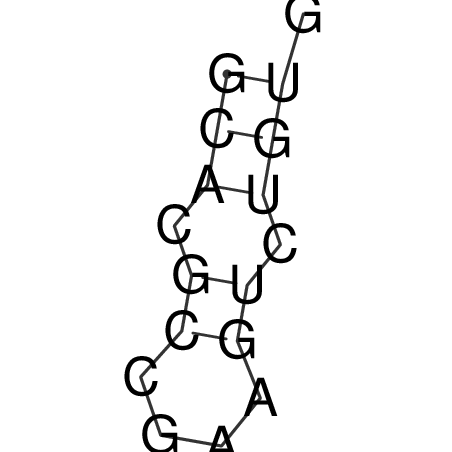
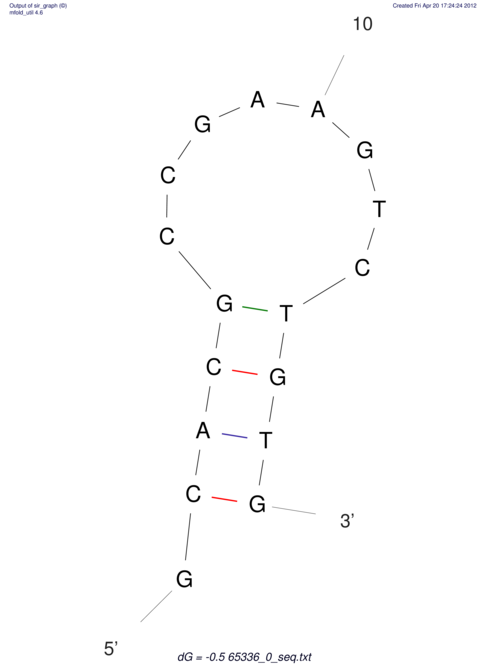
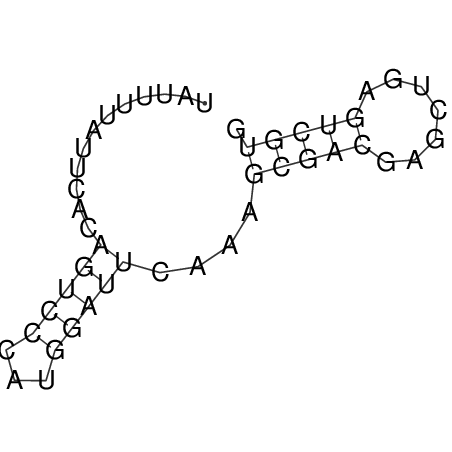
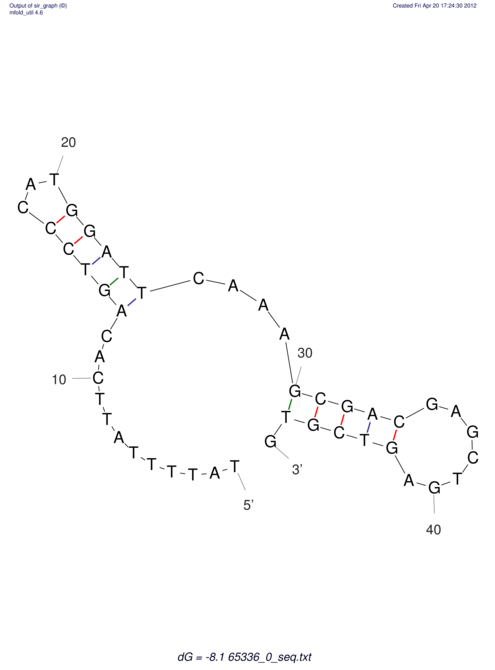
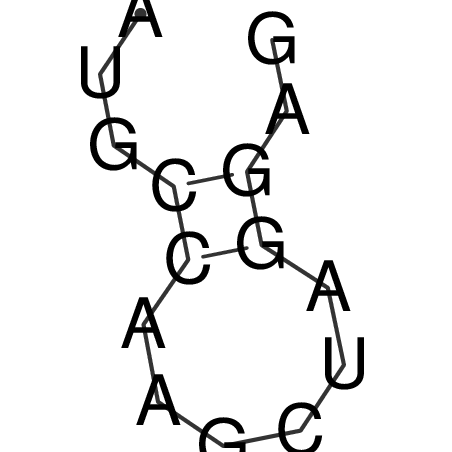
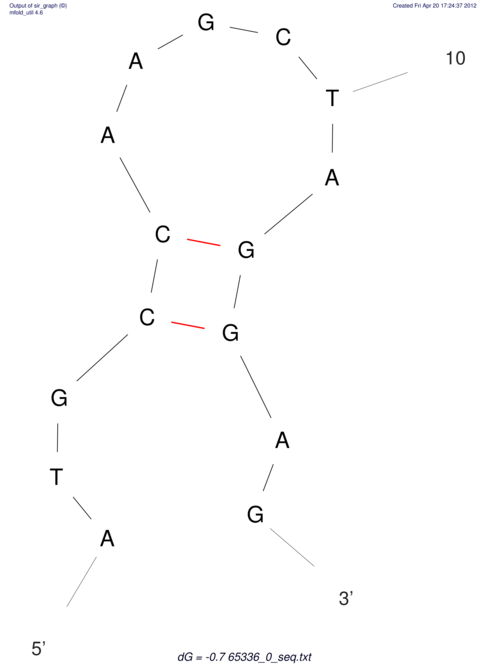
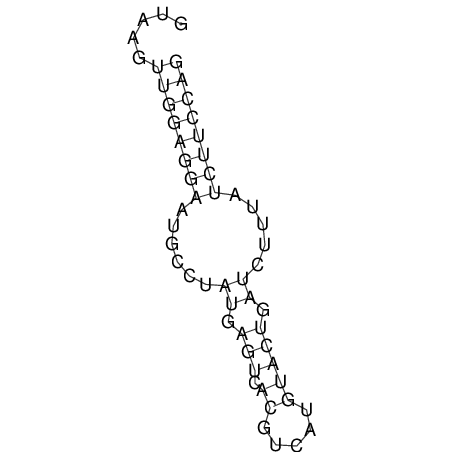
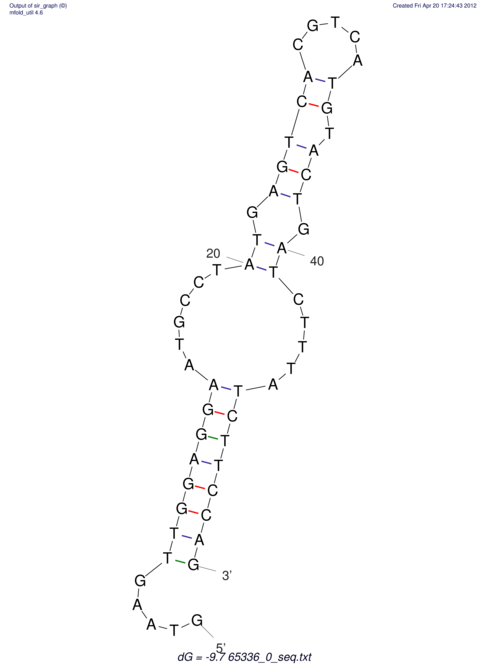
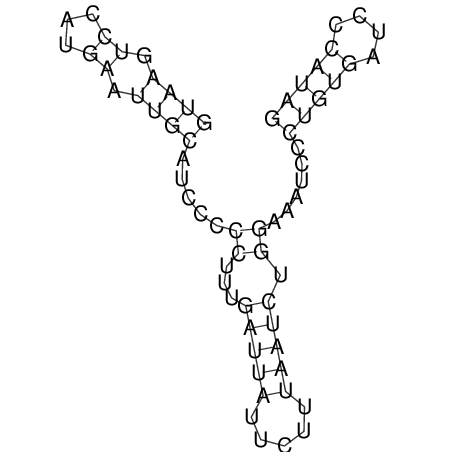
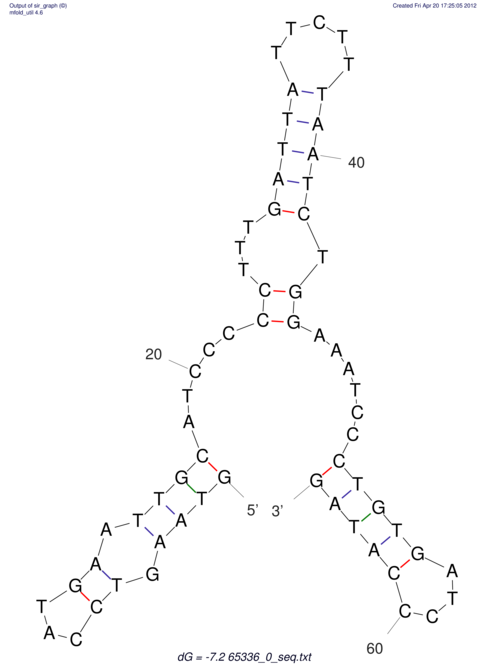
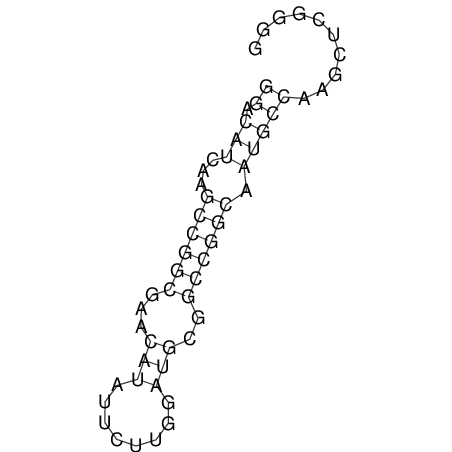
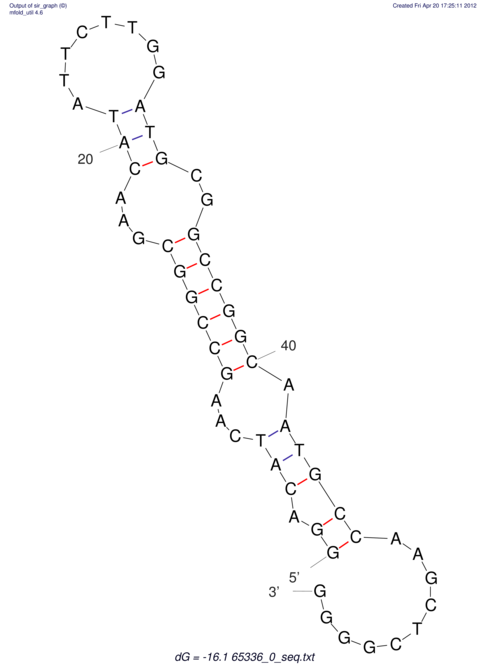
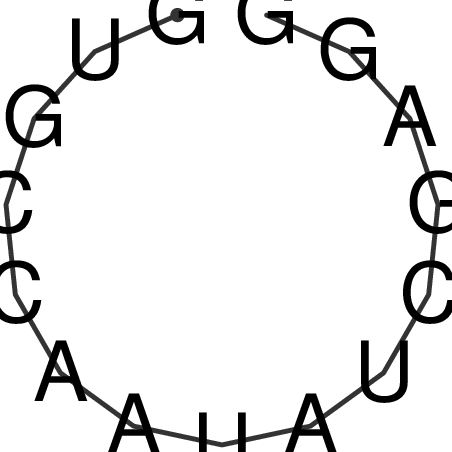
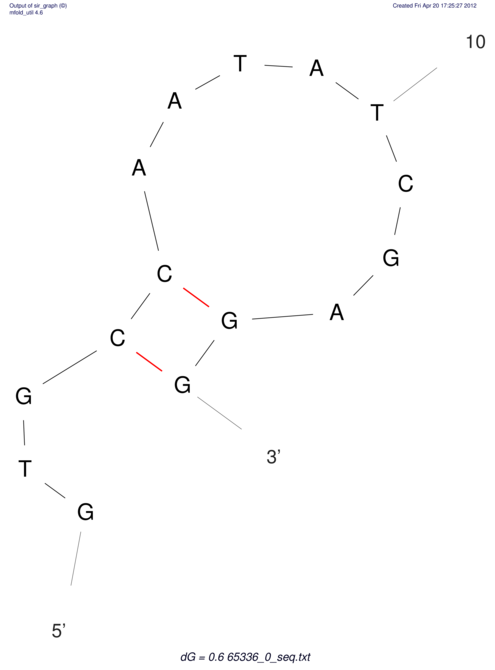
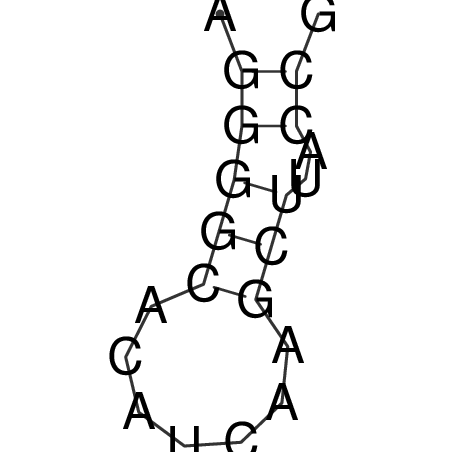
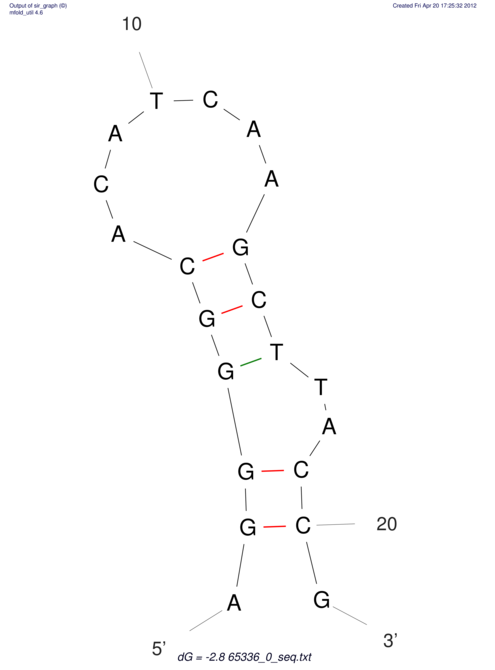
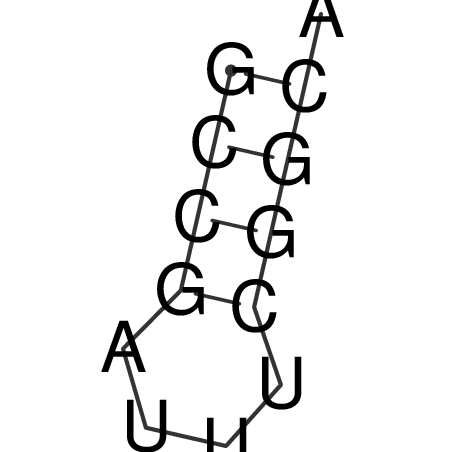
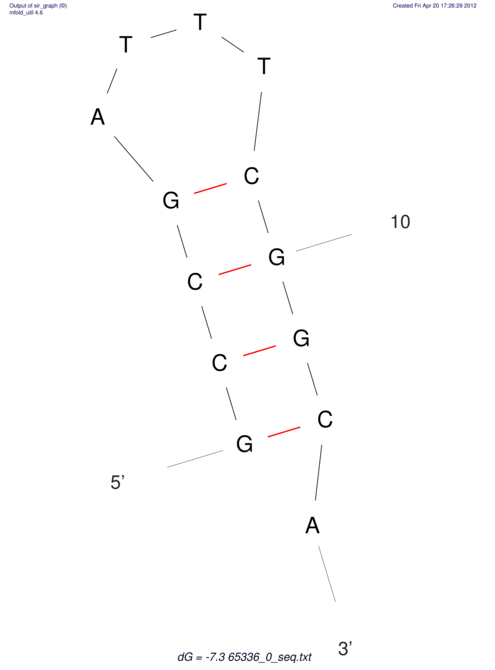
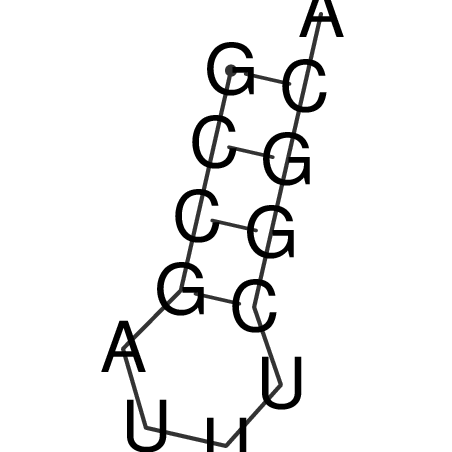
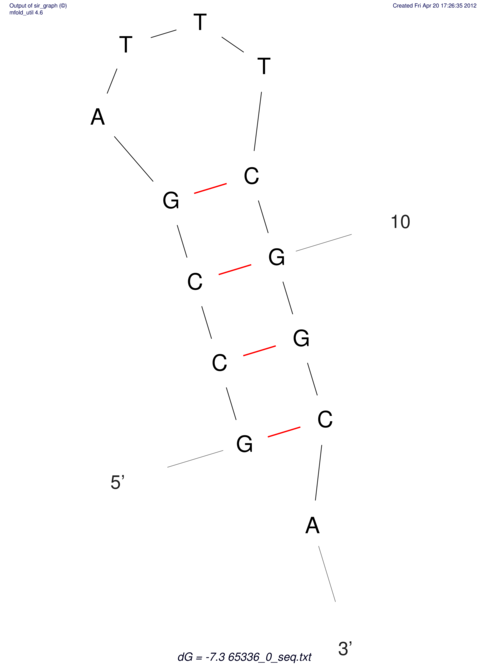
| CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| .....................................................ATAAAAGTATGCCGAACTCG.................. | 20 | 1 | 275.00 | 2.00 | 211.00 | 41.00 | 21.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCA................ | 22 | 1 | 206.00 | 11.00 | 126.00 | 26.00 | 43.00 |
| .....................................................ATAAAAGTATGCCGAACTCGC................. | 21 | 1 | 104.00 | 4.00 | 78.00 | 12.00 | 10.00 |
| ......................................................TAAAAGTATGCCGAACTCGCAGT.............. | 23 | 1 | 100.00 | 4.00 | 18.00 | 10.00 | 68.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCAG............... | 23 | 1 | 78.00 | 48.00 | 12.00 | 2.00 | 16.00 |
| .....................................................ATAAAAGTATGCCGAACTC................... | 19 | 1 | 24.00 | 0.00 | 21.00 | 2.00 | 1.00 |
| ......................................................TAAAAGTATGCCGAACTCGCA................ | 21 | 1 | 22.00 | 1.00 | 14.00 | 4.00 | 3.00 |
| .......................................................AAAAGTATGCCGAACTCGCAGT.............. | 22 | 1 | 20.00 | 0.00 | 5.00 | 1.00 | 14.00 |
| ......................................................TAAAAGTATGCCGAACTCGCAG............... | 22 | 1 | 14.00 | 8.00 | 2.00 | 3.00 | 1.00 |
| ......................................................TAAAAGTATGCCGAACTCGCAGC.............. | 23 | 1 | 13.00 | 0.00 | 3.00 | 2.00 | 8.00 |
| ......................................................TAAAAGTATGCCGAACTCGC................. | 20 | 1 | 12.00 | 0.00 | 8.00 | 3.00 | 1.00 |
| ......................................................TAAAAGTATGCCGAACTCGCAGG.............. | 23 | 1 | 12.00 | 0.00 | 4.00 | 1.00 | 7.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCG................ | 22 | 1 | 11.00 | 0.00 | 4.00 | 4.00 | 3.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCAGT.............. | 24 | 1 | 8.00 | 1.00 | 0.00 | 4.00 | 3.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCT................ | 22 | 1 | 6.00 | 0.00 | 6.00 | 0.00 | 0.00 |
| ..............................TTAATTAATAGGATCGGCC.......................................... | 19 | 1 | 5.00 | 0.00 | 5.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGGATGCCGAACTCGCA................ | 22 | 1 | 4.00 | 1.00 | 2.00 | 0.00 | 1.00 |
| .....................................................ATAAAAGTATGCCGAACT.................... | 18 | 1 | 4.00 | 0.00 | 4.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCAGC.............. | 24 | 1 | 4.00 | 0.00 | 1.00 | 0.00 | 3.00 |
| ........................GTACACTTAATTAATAGGATCGGCC.......................................... | 25 | 1 | 3.00 | 0.00 | 3.00 | 0.00 | 0.00 |
| .............................CTTAATTAATAGGATCGGCC.......................................... | 20 | 1 | 3.00 | 0.00 | 3.00 | 0.00 | 0.00 |
| ....................................AATAGGATCGGCCGTTAATAA.................................. | 21 | 1 | 3.00 | 0.00 | 3.00 | 0.00 | 0.00 |
| .......................................................AAAAGTATGCCGAACTCGCAGG.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| .....................................................ATAAAATTATGCCGAACTCG.................. | 20 | 1 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| ......................................................TAAAATTATGCCGAACTCGC................. | 20 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| .....................................ATAGGATCGGCCGTTAATAAAAGT.............................. | 24 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ............................ACTTAATTAATAGGATCGGCC.......................................... | 21 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ......................................................TAAAAGTATCCCGAACTCGCAGT.............. | 23 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATGCCGAACTGG.................. | 20 | 1 | 2.00 | 0.00 | 0.00 | 2.00 | 0.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCAGG.............. | 24 | 1 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| .....................TTCGTACACTTAATTAATAGGATC.............................................. | 24 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ...................................TAATAGGATCGGCCGTTAATAA.................................. | 22 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .....................................................ATAAAACTATGCCGAACTCGCA................ | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ......................................................TAAAAGTATGCCGAACTCGAC................ | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ......................................................TAAAAGTATGTCGAACTCGCAGT.............. | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ..........................................................AGTATGCCGAACTCGCAGT.............. | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .....................................................ATAAAAGTATACCGATCTCGCAG............... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATGCTGAACTCG.................. | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...............................TAATTAATAGGATCGGCCGTTAA..................................... | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ......................................................TAAAAGTATGCCGAACTC................... | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ......................................................TAAAAGCATGCCGAACTCGCAGT.............. | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................AATAGGATCGGCCGTTAATAAAA................................ | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGAATGCCGAACTCGCAGT.............. | 24 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ....................................................AATAAAAGTATGCCGAACTC................... | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ......................................................TAAAAGTATGCCGAACTCG.................. | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ......................................................TAAAAGTATGCCAAACTCGCAG............... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTTTGCAGAACTCGCAG............... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ...............................TAATTAATAGGATCGGCC.......................................... | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCAC............... | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATGCCGAACTCT.................. | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .................................ATTAATAGGATTGGCCGTTAA..................................... | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTAAGCCGAACTCGC................. | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ......................................................TAAAAGTATGCCGAACTCGCAGTT............. | 24 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .......................................................AAAAGTATGCCGAACTCGCAGC.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................................ATTAATAGGATCGGCCGTTAAT.................................... | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................................ATAACAGTATGCCGAACTCGCAG............... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................ATCAAAGTATGCCGAACTCG.................. | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ....................................................AATAAAAGxATGCCGTACTCGCAG............... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................ATAGGATCGGCCGTTAATAAAAG............................... | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...................................TAATAGGATCGGCCGTTAAT.................................... | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...........................................................GTATGCCGAACTCGCAGT.............. | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCC................ | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ......................................................TAAAAGTATGCCGTACTCGCA................ | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGAATGCCGAACTCG.................. | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................ATAGGATCGGCCGTTAATAAAAGTAT............................ | 26 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .....................................................ATAAAAGTATGCCGAACTCGT................. | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ......................................................TAAAAGTATGCCGAACTCGCT................ | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATGCCGAACTCC.................. | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................ATAAAAGTATGCCGAACTCGCAT............... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATACCGAACTCGCAGT.............. | 24 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................ATAAAAGAATGCCGAACTCGCAG............... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGGATGCCGAACTCGC................. | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTCTGCCGCACTCGCAG............... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATGGCGAACTCG.................. | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ......................................................TAAAAATATACCGAACTCGCA................ | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ....................................................AATAAAAGTATGCCGAACTCGCAGC.............. | 25 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ......................................................TAAAAGTATGCCGGACTCGCAGT.............. | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................AATAGGATCGGCCGTTAATAAAAG............................... | 24 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................ATAGGATCGGCCGTTAATAAAA................................ | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATGCCGAAATCGCAG............... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGGATGCCGAACTCG.................. | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .......................................................AAAAGTATGCCGAACTCGCAG............... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................ATAAAAGTATGTCGAACTCGCA................ | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ......................................TAGGATCGGCCGTTAATAAAAGTATGC.......................... | 27 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
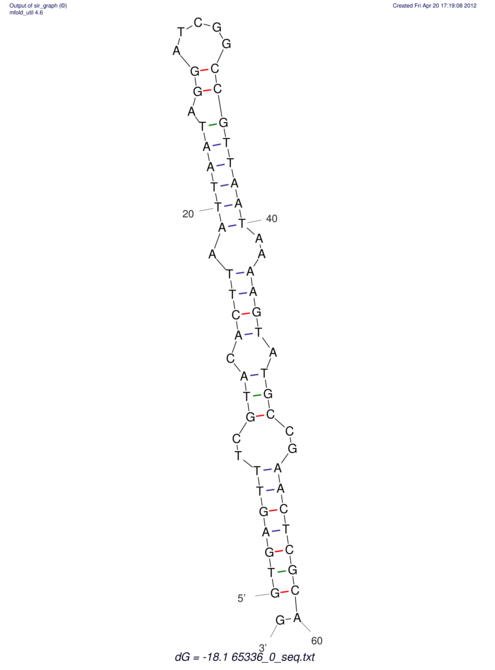
| CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCC---------------------------GAACTCGCAGGCGACGGATGCGCGA TGC--------AGATGCAAGCAGAATGCCCTGAACTAATG-GATTGCCTTTTAATAAAAACAA--CAGATGGATTGAAGCTGTGCGACTTTGGGATCTCGCGGGTGGTG--TGCGAGG | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| ........................................................................ATTGAAGCTGTGCGACTTTG.......................... | 20 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ................................................................................TGCGCGACTTTGGCATCTC................... | 19 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
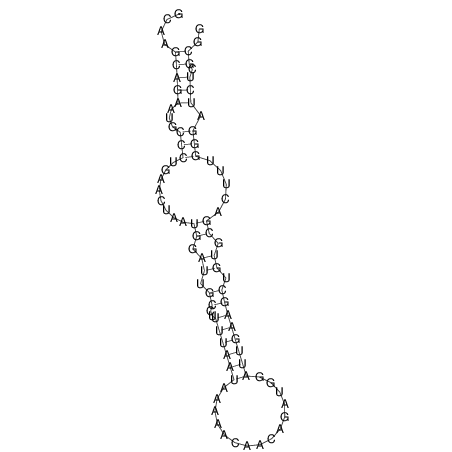
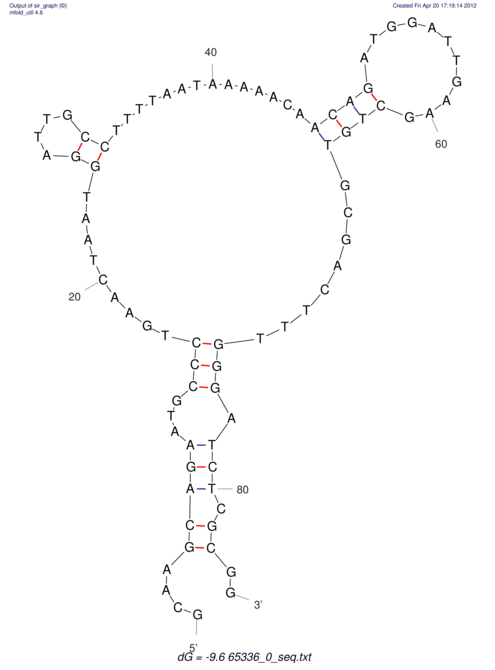
| CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGAC------------GGATGCGCGA AAGCTTGT--------------------AGTTATTAAATAAACTAGCTTGTCC--TGCAGTCCGATGGACTC-CAAGCGACGTGCCGAGTCGTGGAAGCGAAA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
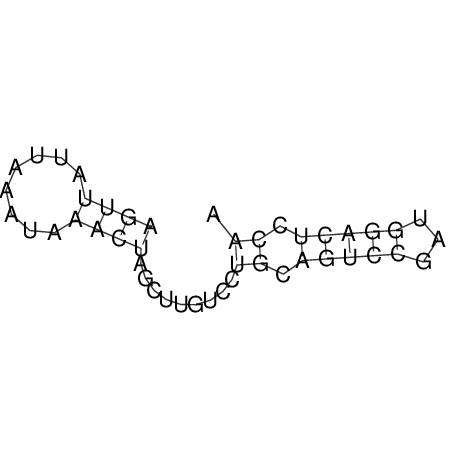
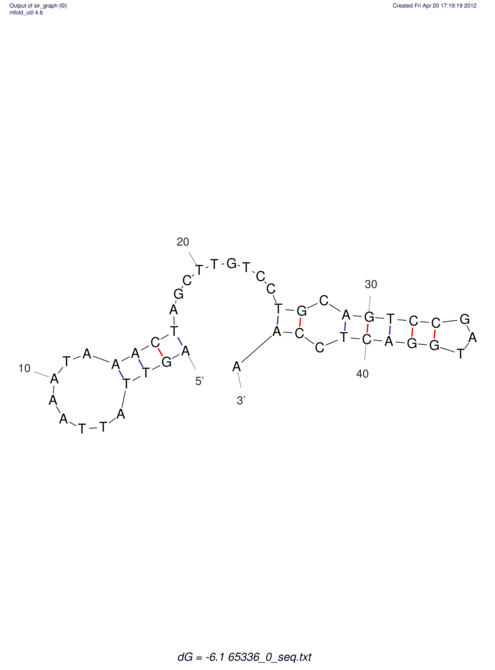
| CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA GTCACTGC-------------------------------------------------AAGCTGGCCGACTTCGGCATGTGCAAGGAGGGCA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| ..CACTGC-------------------------------------------------AAGCGGACCGAC...................... | 18 | 1 | 4.00 | 0.00 | 4.00 | 0.00 | 0.00 |
| .TCACTGC-------------------------------------------------AAGCGGACCGA....................... | 18 | 1 | 3.00 | 0.00 | 3.00 | 0.00 | 0.00 |
| ..............................................................GGCCGACTTCGGCATGTGCAAGGAGGGC. | 28 | 1 | 2.00 | 0.00 | 0.00 | 2.00 | 0.00 |
| .TCACTGC-------------------------------------------------AAGCGGACCGAC...................... | 19 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| .......C-------------------------------------------------AAGCTGGCGGATTTCGGC................ | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ....................................................................CTTCGGCATGAACAAGGA..... | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
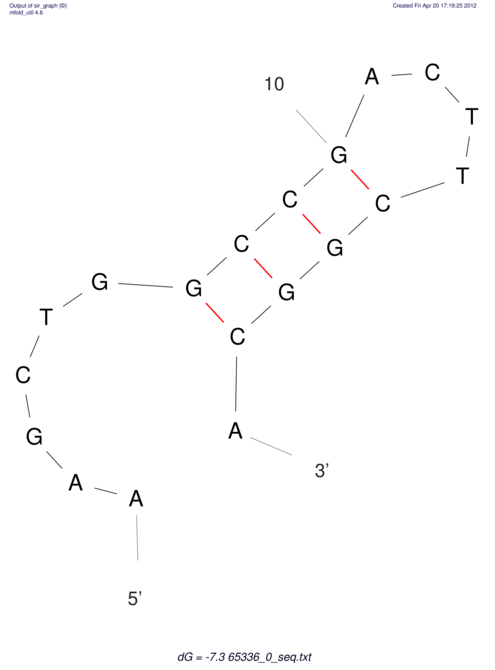
| CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA GCATGTGTAAGGAGG------------------------------------------------------------------------GCAT | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA C--------------------------------------------------------------------------------GAATGGGCGC | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA GGCTACGCCCG---------------------------------------------------------------------------GATCA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |
| CACCGTGCTGGAGAGGTGAGTTTCGTACACTTAATTAATAGGATCGGCCGTTAATAAAAGTATGCCGAACTCGCAGGCGACGGATGCGCGA C--------------------------------------------------------------------------------------GCGA | Size | Hit Count | Total Norm | V044 Embryo | M023 Head | M024 Male body | M025 Embryo |