
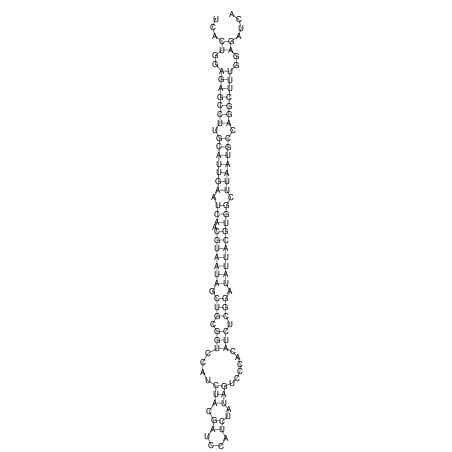
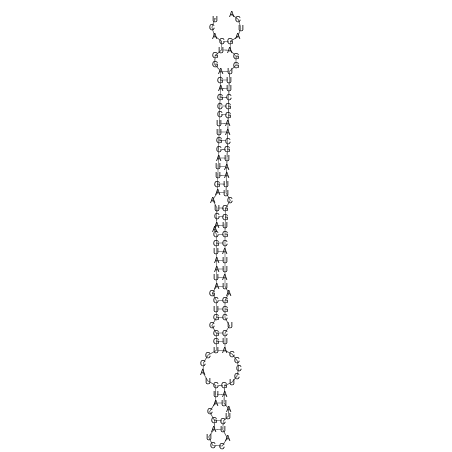
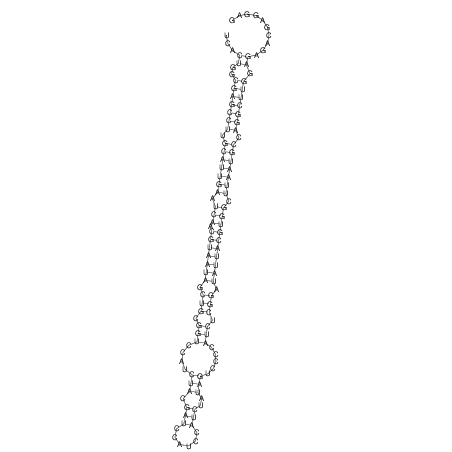
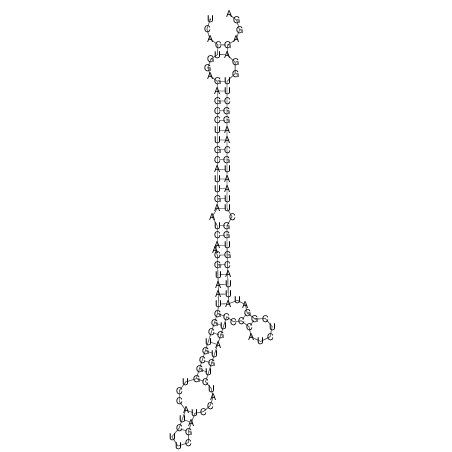
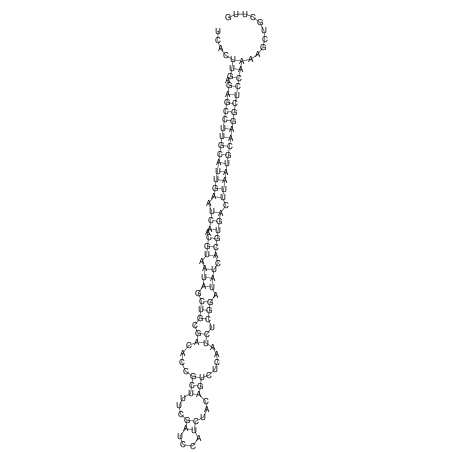
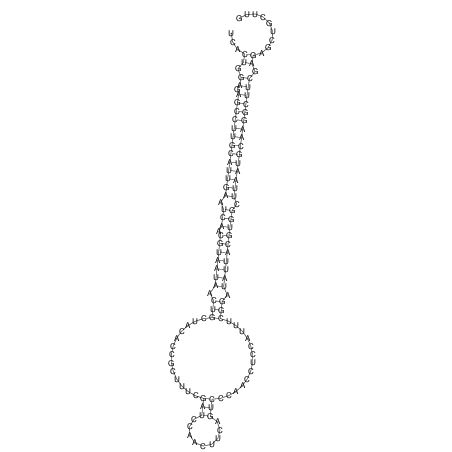
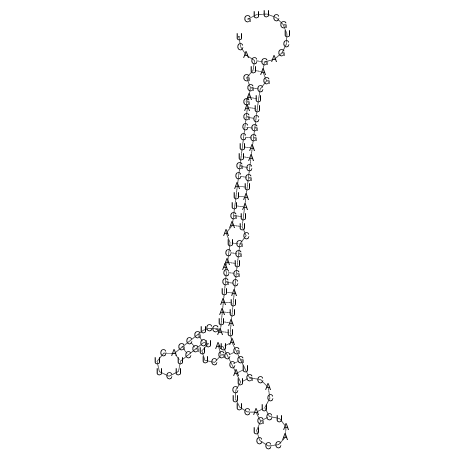

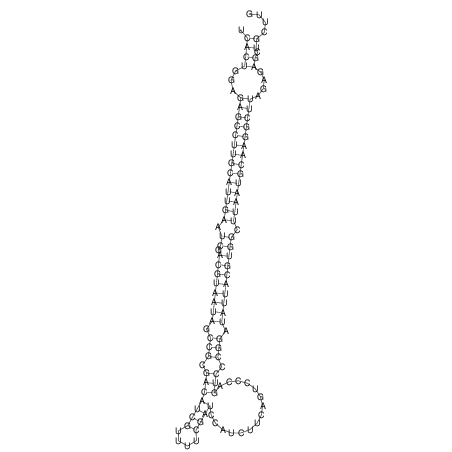


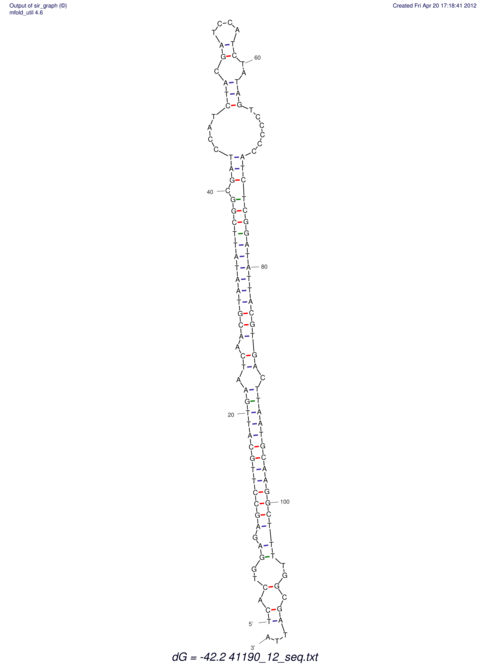
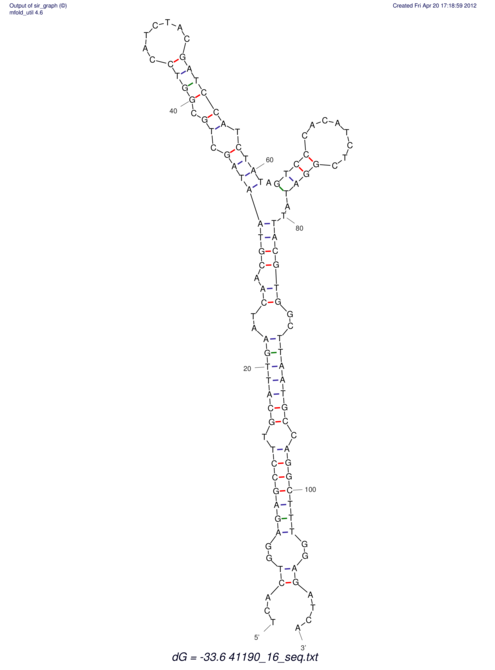
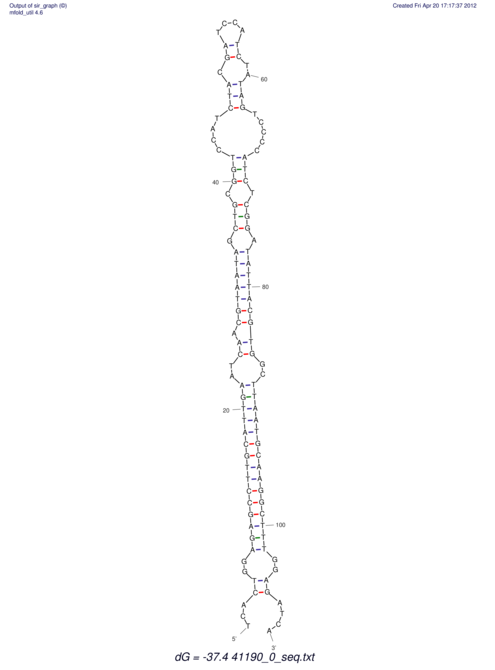
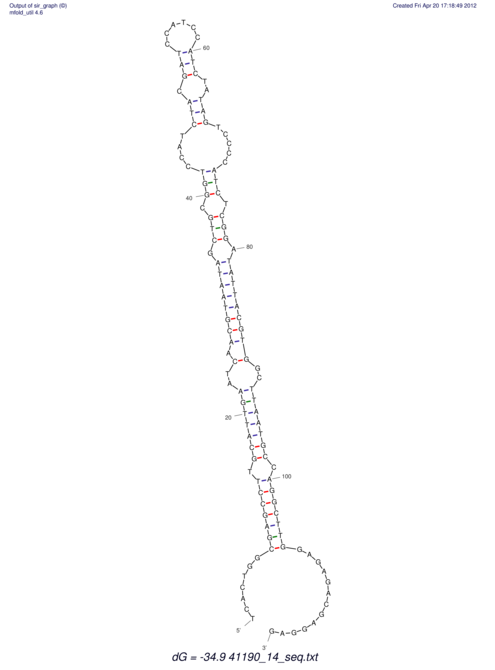
View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
block92371 |
chr2L:6973236-6973348 - |
Candidate |
|---|---|---|
| [View on UCSC Genome Browser {Cornell Mirror}] | three_prime_UTR |
Legend:
mature strand at position 6973248-6973268| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr2L:6973221-6973363 - | 1 | CGCGCCGAAGAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATATTC-GGCGAT-------------CC-ATCTACG-ATC----C-ATCTATAGTCCCC--CATC--------T---CGGATATTACGTGACTTAATGCAAGGCTTTTGGC---------------------------------------------------------------GATTAAAGTCAAG-----------------------CGGAG-------AT | |
| droSim1 | chr2L:6769566-6769705 - | LASTZ | 4 | CC--CCGAAGAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATAGCT--GCGGT-------------CC-ATCTACG-ATC----C-ATCTATAGTCCCA--CATC--------T---CGGATATTACGTGGCTTAATGCCAGGCTTT-GGA----------------------------------------------------------G-----ATCAAAGTCGAG-----------------------CGGAG-------AT |
| droSec1 | super_3:2505870-2506008 - | LASTZ | 1 | CC--CCGAAGAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATAGCT--GCGGT-------------CC-ATCTACG-ATC----C-ATCTATAGTCCCC---ATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTTT-GGA----------------------------------------------------------G-----ATCAAAGTCGAG-----------------------CGGAG-------AT |
| droYak2 | chr2L:5936133-5936280 + | LASTZ | 3 | CC---CCAAGAATACTCACT------------GG----CGAGCCTTGCATTGAATCAACGTAATAGCT--GCGGT-------------CC-ATCTACG-ATCCATCC-ATCTATAGTCCCC---ATC--------T---CGGATATTACGTGGCTTAATGCCAGGCTTG-G-AGAGA-CGAG----------------------------------------------GAG----------GAGTCGAG-----------------------CAGAG-----ACGC |
| droEre2 | scaffold_4929:15889020-15889155 - | LASTZ | 3 | CC----CAAGAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATGGCT--GCGGT-------------CC-ATCTTCG-ATC----C-ATCTGTAGTCCCC---ATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTTG-G-A----------------------------------------------------------GAG-----GAAAGTCGAG-----------------------AGGAG-------AC |
| droEug1 | scf7180000409463:94621-94763 - | LASTZ | 4 | CC----CACAAATACTCACT------------TG----AGAGCCTTGCATTGAATCAACGTAATAGCT--GCGAC------------ACC-GCTTTCG-ATC----C-ATCTACAGTCTCA---ATC--------T---CGGATATCACGTGACTTAATGCAAGGCTCC-A-A----------------------------------------------------------AAGCTGCTTGAAAATGCG---------------------AGTCGAG-------AA |
| droBia1 | scf7180000302422:2439201-2439346 + | LASTZ | 4 | CC-----AAAAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATAACT--GCTAC------------ACC-GCTTTCG-ATC----C-AACTTCAGTCCCA---ACCTCCA--TTT---CGGATATTACGTGGCTTAATGCAAGGCTTC-G-A----------------------------------------------------------GAGCTGCTTGAAAACGCG-----------------------AGGCA-------AG |
| droTak1 | scf7180000416008:231160-231307 - | LASTZ | 4 | CC----CACAAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATAGCT--GCGAC-------TTC--TTC-GCTTTCG-ATC----C-ATCTTCAGTCCCA---ATCTC-------ACGTGGATATTACGTGGCTTAATGCAAGGCTTC-G-A----------------------------------------------------------GAGCTGCTTGAAAACGCG-----------------------AGGCG-------AG |
| droEle1 | scf7180000490640:333648-333793 + | LASTZ | 4 | CC-----ACAAATACTCACT------------GG----AGAGCCTTGTATTGAATCAACGTAATAGCT--GCGAT------CGTCTAATC-GTCTTCG-ATC----C-ATCTTCAGTCCCA---ATC--------G---CGGATATTACGTGGCTTAATGCAAGGCTTT-G-A----------------------------------------------------------GAGCTGATTGAAAACGCG-----------------------ATACG-------AT |
| droRho1 | scf7180000779930:68908-69048 - | LASTZ | 4 | CC-----ACAAATACTCACT------------GG----AGAGCCTTGCATTGAATCGACGTAATAGCC--GCGAC------------ATC-GTTTTCG-ATC----C-ATCTTCAGTCCCA---GTC--------C---CGGATATTACGTGGCTTAATGCAAGGCTTA-G-A----------------------------------------------------------GAGCTGCTTGAA-TCGCG---------------------ATACGAG-------AC |
| droFic1 | scf7180000452011:146-285 - | LASTZ | 4 | CC-----AAAAATACTCACT------------GG----GGAGCCTTGCATTGAATCAACGTAATAGCT--GCGAC------------CTC-GTTTTCG-ATC----C-ATCTTTAGTCCCA---ATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTTC-G-G----------------------------------------------------------GAGCCGATTGAAGTCGAG-----------------------ACGAG-------AG |
| droKik1 | scf7180000302468:483720-483875 - | LASTZ | 4 | CC----CACAAATACTCACT------------GG----AGAGCCTTGCATTGAATCGACGTAATGGTCTCGCGAC-------------AC-GATTTCG-AAC----C-ATCTTCAGTCCCAAATATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTAC-G-A----------------------------------------------------------GAGCTCCTTCAGAACGCG----AC--------AA-AACGTAACGAG-------AC |
| droAna3 | scaffold_12916:53076-53242 - | LASTZ | 1 | GGCGCTCACAAATACTCACT------------GTAA--TGAGCCTTGCATTGAATCAACGTAATAGCT--GTGTC-------GGC---TCTATTTTGG-TCC----G-AAATTCAGTCCCA---ATC--------T---GGGATATTACGTGGCTTAATGTAAGGCTTT-G-G----------------------------------------------------------GAGCTGCTTGAAGAAATA--AATTTA------AA-AAA----GGAGAAAGACCTT |
| droBip1 | scf7180000392937:103419-103594 - | LASTZ | 4 | GGCGCCCACAAATACTCACT------------GAAT--AGAGCCTTGCATTGAATCAACGTAATAGCT--GTGAC-------AGC---TC-TATTCCG-GTC----CAAACTTCAGTCCCA---ATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTTT-G-G----------------------------------------------------------GAGCTGCTCGAAAAAAAATTAAAATAACTTTAAAGAAA--AACGCA--AGACCTT |
| dp4 | chr4_group1:3766092-3766263 + | LASTZ | 3 | TGCCCCCCAAAATACTCATT------------GGAGGGAGAGCCTTGCATTGAATCAACGTAATTGCT--GTGAACAAAATCATCTA-TC-GAATCCGAGTC----CAAATGTCAG--CCA---ATT--------ACAGCAGCAATTACGTGGCTTAATGCCAGGCTTT-A-AGAGAGAG------------------------------------------------ACAGAGCTGCTCGAAA------------------------CGAAGCGCA-------AA |
| droPer1 | super_5:1509790-1509963 - | LASTZ | 4 | TGCCCCCCAAAATACTCATT------------GGAGGGAGAGCCTTGCATTGAATCAACGTAATTGCT--GTGAACAAAATCATCTA-TC-GAATCCGAGTC----CAAATGTCAGC--CA---ATA--------ACAGCAGCAATTACGTGGCTTAATGCCAGGCTTT-A-AGAGAGAGAG----------------------------------------------AGAGAGCTGCTCGAAA------------------------CGAAGCGCA-------AA |
| droWil1 | scaffold_180772:7270846-7270981 + | LASTZ | 1 | TGAACACAAAAAT---AAAA------------AG----TTAGCCTAGCATTGAATCGACGTAATAGTT--TGAGT-------------TC-AT---AA-ATC----C-ATT-------GAA---ACT--------T---TGACAATTACGTGACTTAATGCTAAGCTTT-G-T----------------------------------------------------------AA-CAAAATGAA----------AATA------GA-AAAGAAACAAG-------AT |
| droVir3 | scaffold_12963:3413645-3413810 - | LASTZ | 3 | CCAAAAAAAAAATACTCATTGAAAGCGT-GTTTG----AAAGCTTTGCATTGAATCGCAGTAATCGGCT-GGATT-CACAGAAA--------CCTTAG-AAC----T-CT--ATTATAACT---TTT---TA-AATCAGCTGCAATTACGTGGCTTAATGCAAAGCTTG-G-AATGTGAGGA----------------------------------------------AAAAAGCAGCTTT---------------------------------AT---------G |
| droMoj3 | scaffold_6500:13787240-13787449 - | LASTZ | 3 | AGAATCATAAAATACTCATTGAGAGCGTTATGCA----AAAGCTTTGCATTGAATCGCCGTAATCGCA--GT--C------------CAC-ACTTTAG-ATA----T-TTT-TTTGTAACC---TTC---GAAAATCAGCTGCAATTACGCGGCTTAATGCAAAGCTTA-T-GATGCAAGAAAAAAAAGAACGAAATATTAATGATAAAATAACAATAAAAAATTAAAAGAAAGCTGCTTT---------------------------------ATG-------AA |
| droGri2 | scaffold_15252:2977193-2977325 - | LASTZ | 4 | CA----TTAAAAGCTTCATT------------TG----AAAGCTTTGCATTGAATCGCAGTAATCTCG-AGCTAC------------ATA-TCCTTAG-AAC----T-TT--------------ATT--------GCTGTTTTAATTACGTGGCTTAATGCAAAGCTTG-A-AATAAACGA-------------------------------------------------AAAGCTGCTTT---------------------------------ATG-------AA |
| Species | Alignment |
| dm3 | CGCGCCGAAGAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATATTC-GGCGAT-------------CC-ATCTACG-ATC----C-ATCTATAGTCCCC--CATC--------T---CGGATATTACGTGACTTAATGCAAGGCTTTTGGC---------------------------------------------------------------GATTAAAGTCAAG-----------------------CGGAG-------AT |
| droSim1 | CC--CCGAAGAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATAGCT--GCGGT-------------CC-ATCTACG-ATC----C-ATCTATAGTCCCA--CATC--------T---CGGATATTACGTGGCTTAATGCCAGGCTTT-GGA----------------------------------------------------------G-----ATCAAAGTCGAG-----------------------CGGAG-------AT |
| droSec1 | CC--CCGAAGAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATAGCT--GCGGT-------------CC-ATCTACG-ATC----C-ATCTATAGTCCCC---ATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTTT-GGA----------------------------------------------------------G-----ATCAAAGTCGAG-----------------------CGGAG-------AT |
| droYak2 | CC---CCAAGAATACTCACT------------GG----CGAGCCTTGCATTGAATCAACGTAATAGCT--GCGGT-------------CC-ATCTACG-ATCCATCC-ATCTATAGTCCCC---ATC--------T---CGGATATTACGTGGCTTAATGCCAGGCTTG-G-AGAGA-CGAG----------------------------------------------GAG----------GAGTCGAG-----------------------CAGAG-----ACGC |
| droEre2 | CC----CAAGAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATGGCT--GCGGT-------------CC-ATCTTCG-ATC----C-ATCTGTAGTCCCC---ATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTTG-G-A----------------------------------------------------------GAG-----GAAAGTCGAG-----------------------AGGAG-------AC |
| droEug1 | CC----CACAAATACTCACT------------TG----AGAGCCTTGCATTGAATCAACGTAATAGCT--GCGAC------------ACC-GCTTTCG-ATC----C-ATCTACAGTCTCA---ATC--------T---CGGATATCACGTGACTTAATGCAAGGCTCC-A-A----------------------------------------------------------AAGCTGCTTGAAAATGCG---------------------AGTCGAG-------AA |
| droBia1 | CC-----AAAAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATAACT--GCTAC------------ACC-GCTTTCG-ATC----C-AACTTCAGTCCCA---ACCTCCA--TTT---CGGATATTACGTGGCTTAATGCAAGGCTTC-G-A----------------------------------------------------------GAGCTGCTTGAAAACGCG-----------------------AGGCA-------AG |
| droTak1 | CC----CACAAATACTCACT------------GG----AGAGCCTTGCATTGAATCAACGTAATAGCT--GCGAC-------TTC--TTC-GCTTTCG-ATC----C-ATCTTCAGTCCCA---ATCTC-------ACGTGGATATTACGTGGCTTAATGCAAGGCTTC-G-A----------------------------------------------------------GAGCTGCTTGAAAACGCG-----------------------AGGCG-------AG |
| droEle1 | CC-----ACAAATACTCACT------------GG----AGAGCCTTGTATTGAATCAACGTAATAGCT--GCGAT------CGTCTAATC-GTCTTCG-ATC----C-ATCTTCAGTCCCA---ATC--------G---CGGATATTACGTGGCTTAATGCAAGGCTTT-G-A----------------------------------------------------------GAGCTGATTGAAAACGCG-----------------------ATACG-------AT |
| droRho1 | CC-----ACAAATACTCACT------------GG----AGAGCCTTGCATTGAATCGACGTAATAGCC--GCGAC------------ATC-GTTTTCG-ATC----C-ATCTTCAGTCCCA---GTC--------C---CGGATATTACGTGGCTTAATGCAAGGCTTA-G-A----------------------------------------------------------GAGCTGCTTGAA-TCGCG---------------------ATACGAG-------AC |
| droFic1 | CC-----AAAAATACTCACT------------GG----GGAGCCTTGCATTGAATCAACGTAATAGCT--GCGAC------------CTC-GTTTTCG-ATC----C-ATCTTTAGTCCCA---ATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTTC-G-G----------------------------------------------------------GAGCCGATTGAAGTCGAG-----------------------ACGAG-------AG |
| droKik1 | CC----CACAAATACTCACT------------GG----AGAGCCTTGCATTGAATCGACGTAATGGTCTCGCGAC-------------AC-GATTTCG-AAC----C-ATCTTCAGTCCCAAATATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTAC-G-A----------------------------------------------------------GAGCTCCTTCAGAACGCG----AC--------AA-AACGTAACGAG-------AC |
| droAna3 | GGCGCTCACAAATACTCACT------------GTAA--TGAGCCTTGCATTGAATCAACGTAATAGCT--GTGTC-------GGC---TCTATTTTGG-TCC----G-AAATTCAGTCCCA---ATC--------T---GGGATATTACGTGGCTTAATGTAAGGCTTT-G-G----------------------------------------------------------GAGCTGCTTGAAGAAATA--AATTTA------AA-AAA----GGAGAAAGACCTT |
| droBip1 | GGCGCCCACAAATACTCACT------------GAAT--AGAGCCTTGCATTGAATCAACGTAATAGCT--GTGAC-------AGC---TC-TATTCCG-GTC----CAAACTTCAGTCCCA---ATC--------T---CGGATATTACGTGGCTTAATGCAAGGCTTT-G-G----------------------------------------------------------GAGCTGCTCGAAAAAAAATTAAAATAACTTTAAAGAAA--AACGCA--AGACCTT |
| dp4 | TGCCCCCCAAAATACTCATT------------GGAGGGAGAGCCTTGCATTGAATCAACGTAATTGCT--GTGAACAAAATCATCTA-TC-GAATCCGAGTC----CAAATGTCAG--CCA---ATT--------ACAGCAGCAATTACGTGGCTTAATGCCAGGCTTT-A-AGAGAGAG------------------------------------------------ACAGAGCTGCTCGAAA------------------------CGAAGCGCA-------AA |
| droPer1 | TGCCCCCCAAAATACTCATT------------GGAGGGAGAGCCTTGCATTGAATCAACGTAATTGCT--GTGAACAAAATCATCTA-TC-GAATCCGAGTC----CAAATGTCAGC--CA---ATA--------ACAGCAGCAATTACGTGGCTTAATGCCAGGCTTT-A-AGAGAGAGAG----------------------------------------------AGAGAGCTGCTCGAAA------------------------CGAAGCGCA-------AA |
| droWil1 | TGAACACAAAAAT---AAAA------------AG----TTAGCCTAGCATTGAATCGACGTAATAGTT--TGAGT-------------TC-AT---AA-ATC----C-ATT-------GAA---ACT--------T---TGACAATTACGTGACTTAATGCTAAGCTTT-G-T----------------------------------------------------------AA-CAAAATGAA----------AATA------GA-AAAGAAACAAG-------AT |
| droVir3 | CCAAAAAAAAAATACTCATTGAAAGCGT-GTTTG----AAAGCTTTGCATTGAATCGCAGTAATCGGCT-GGATT-CACAGAAA--------CCTTAG-AAC----T-CT--ATTATAACT---TTT---TA-AATCAGCTGCAATTACGTGGCTTAATGCAAAGCTTG-G-AATGTGAGGA----------------------------------------------AAAAAGCAGCTTT---------------------------------AT---------G |
| droMoj3 | AGAATCATAAAATACTCATTGAGAGCGTTATGCA----AAAGCTTTGCATTGAATCGCCGTAATCGCA--GT--C------------CAC-ACTTTAG-ATA----T-TTT-TTTGTAACC---TTC---GAAAATCAGCTGCAATTACGCGGCTTAATGCAAAGCTTA-T-GATGCAAGAAAAAAAAGAACGAAATATTAATGATAAAATAACAATAAAAAATTAAAAGAAAGCTGCTTT---------------------------------ATG-------AA |
| droGri2 | CA----TTAAAAGCTTCATT------------TG----AAAGCTTTGCATTGAATCGCAGTAATCTCG-AGCTAC------------ATA-TCCTTAG-AAC----T-TT--------------ATT--------GCTGTTTTAATTACGTGGCTTAATGCAAAGCTTG-A-AATAAACGA-------------------------------------------------AAAGCTGCTTT---------------------------------ATG-------AA |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
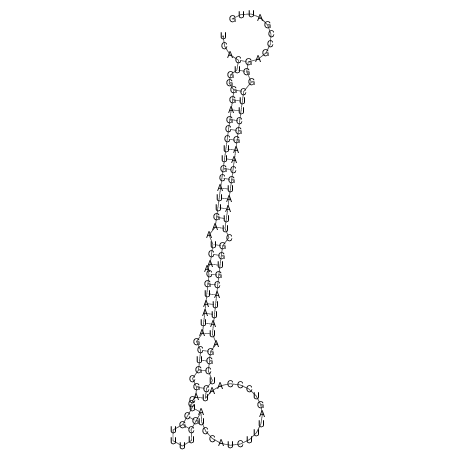
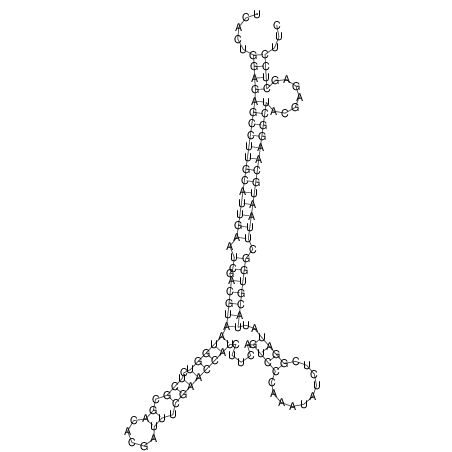
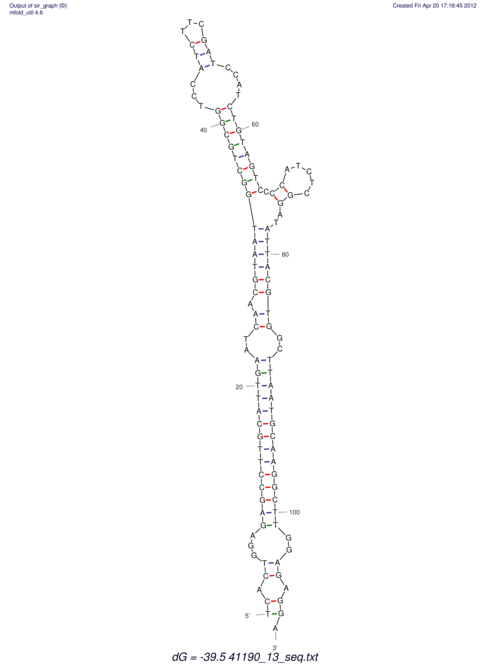
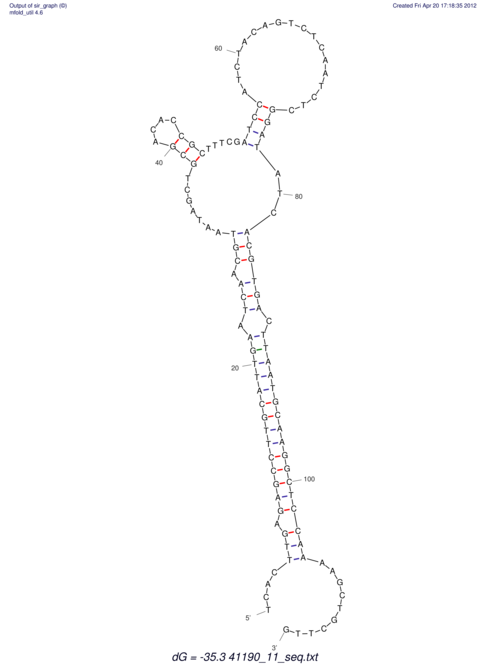
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
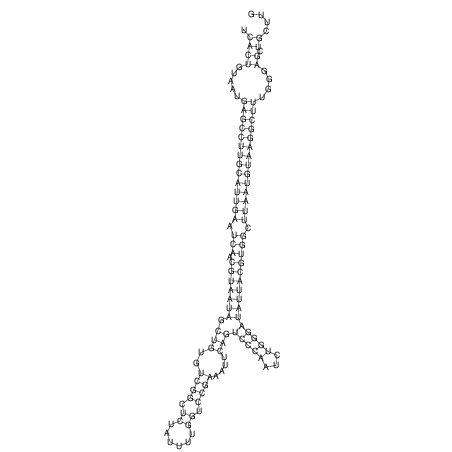
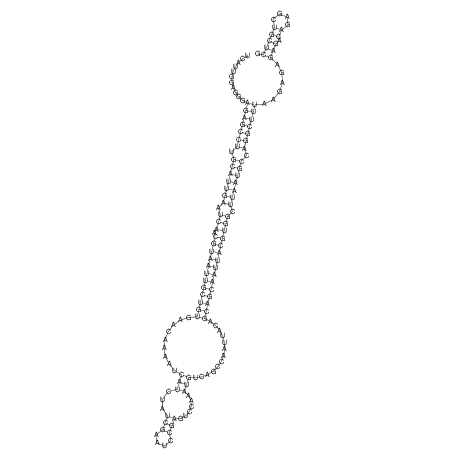
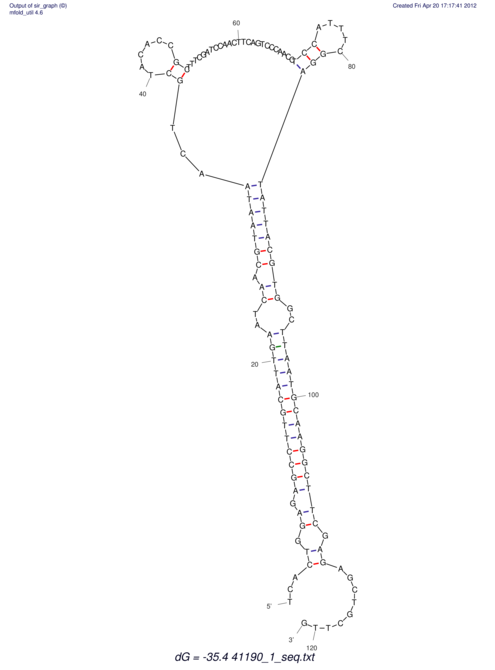
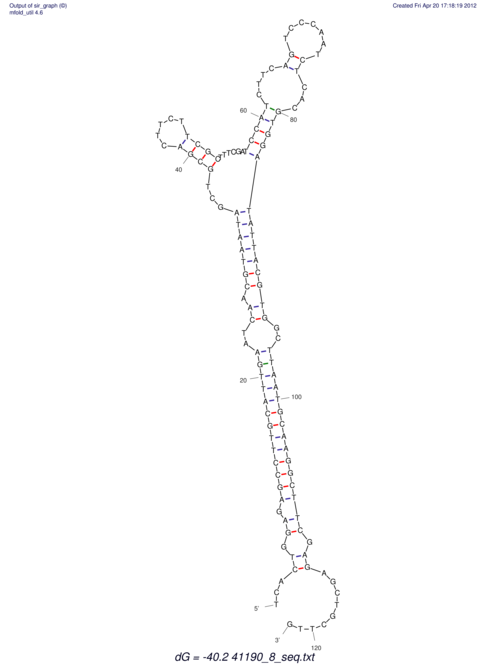
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:19:17 EDT