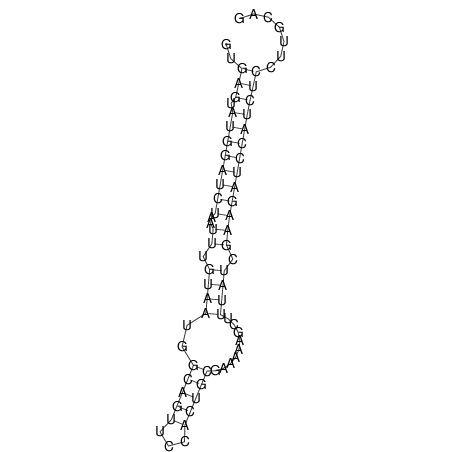
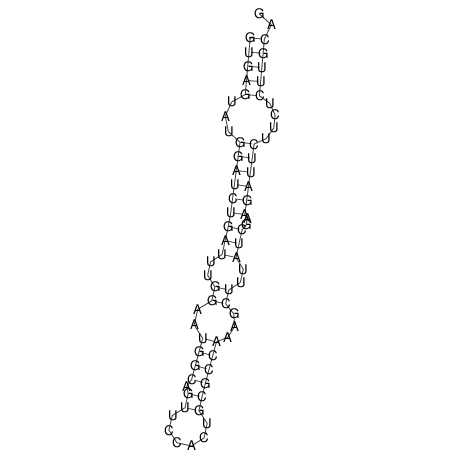
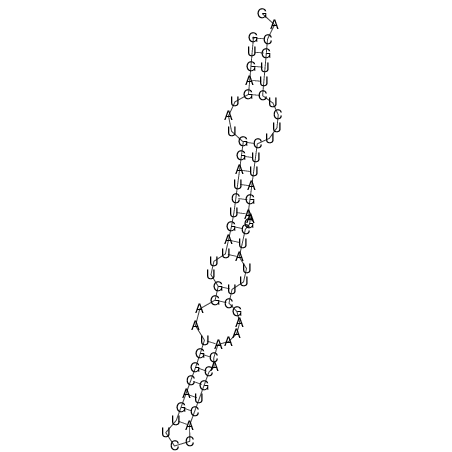
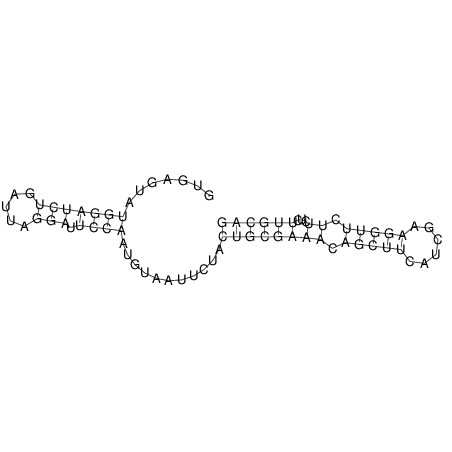
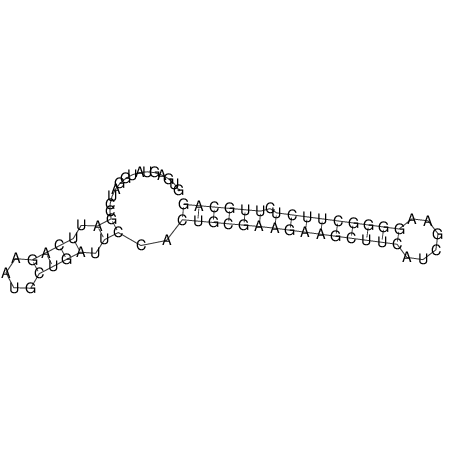
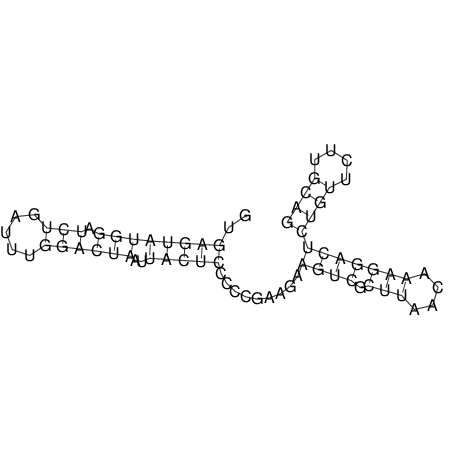
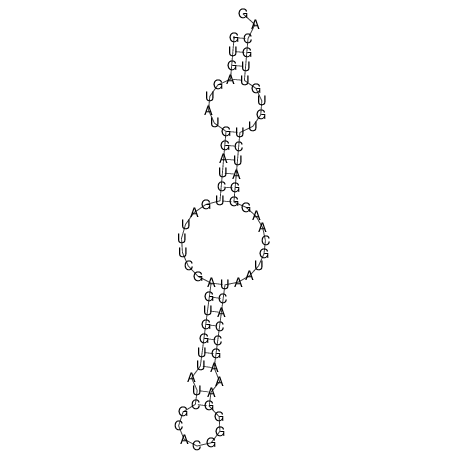
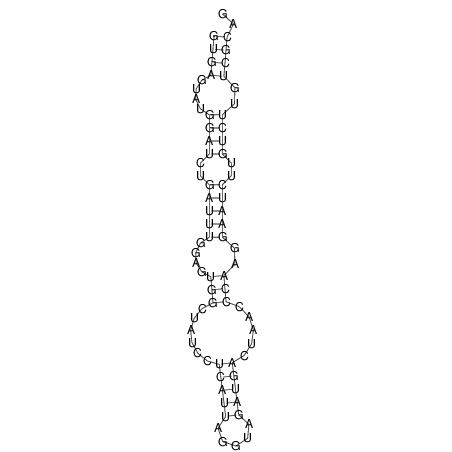



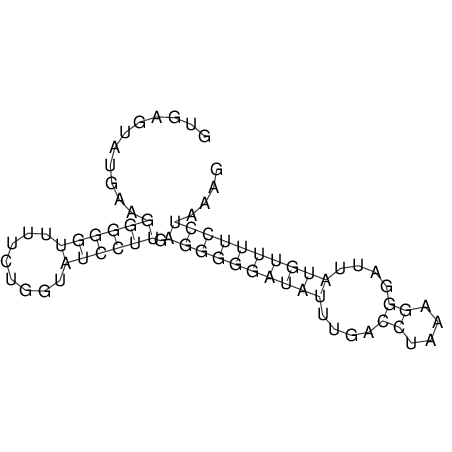
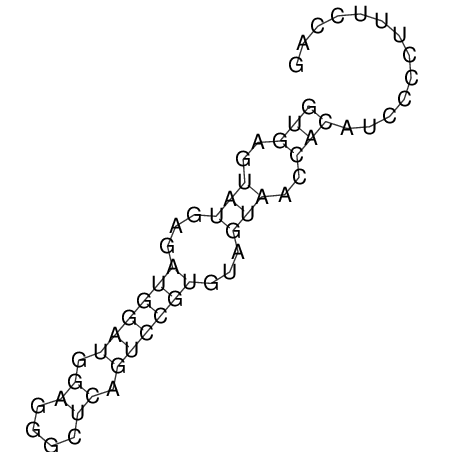
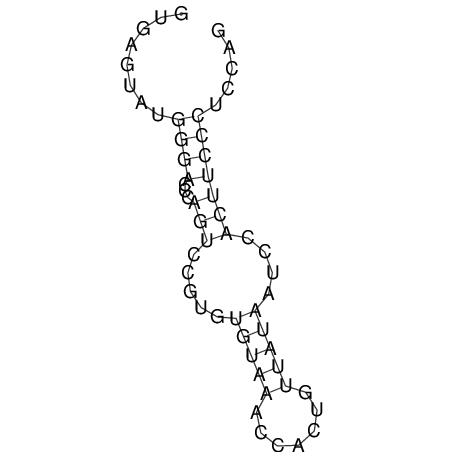

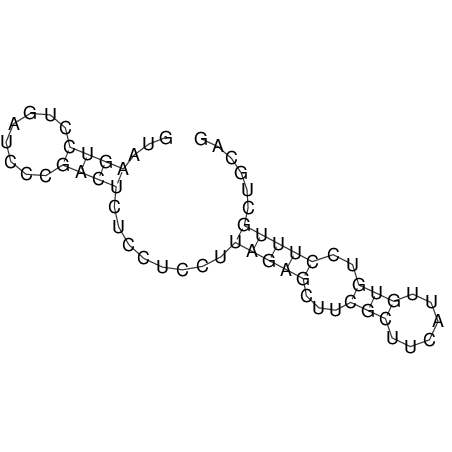
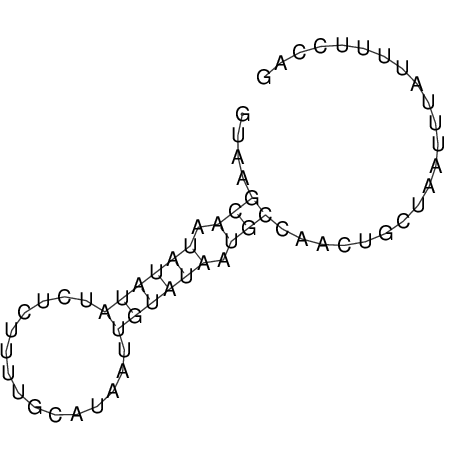
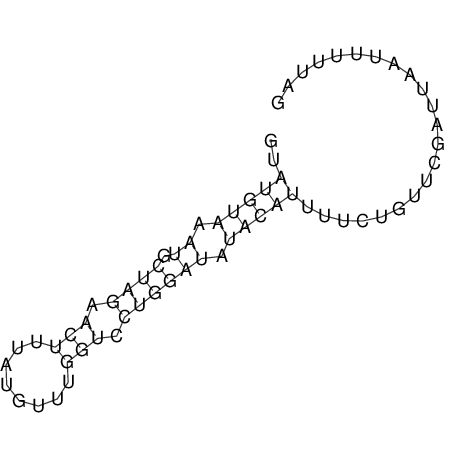
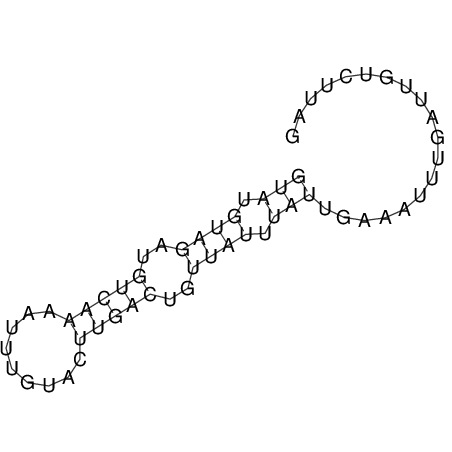
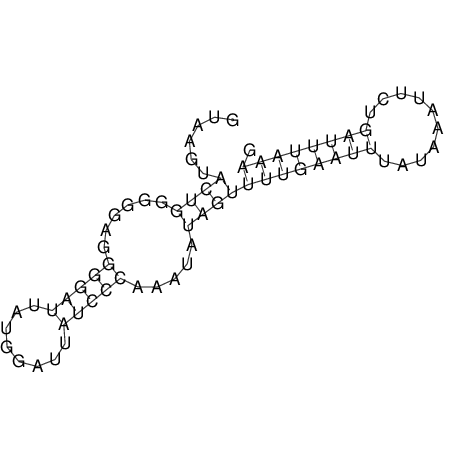



View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
Eaat2_in6 |
chr2L:738436-738507 - |
Candidate |
|---|---|---|
| [View on UCSC Genome Browser {Cornell Mirror}] | mirtron |
Legend:
mature strand at position 738487-738507| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr2L:738421-738522 - | 1 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTAATTTG-----------TAATGGCAGT-----TC-CACT-GCGAA--AAAGCTTTATCGAA-G--------------ATCCAT---CTCCTTGCAGGGATCGGATTCGCAC | |
| droSim1 | chr2L:739962-740062 - | LASTZ | 3 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GAATGGCAGT-----TC-CACT-GCGCC--AAAGCTTTATCGAA----------------GATTCT---TCTCTTGCAGGGATCGGATTCGCAC |
| droSec1 | super_14:710120-710220 - | LASTZ | 4 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GAATGGCAGT-----TC-CACT-GCACA--AAAGCTTTATCGAA----------------GATTCT---TCTCTTGCAGGGATCGGATTCGCAC |
| droYak2 | chr2L:721660-721764 - | LASTZ | 5 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTGATTAG-----------GATTCC-AATGTAATTC-TACT-GCGAA--ACAGCTTCATCGAA----------------GGTTCT---TCTCTTGCAGGGATCGGATTCGCAC |
| droEre2 | scaffold_4929:793076-793176 - | LASTZ | 5 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CCGATTCA-----------GAATGCTGAT-----TC-CACT-GCGAA--GAAGCTTCATCGAA----------------GGGGCT---TCTCTTGCAGGGATCGGATTCGCAC |
| droEug1 | scf7180000409554:1463548-1463649 - | LASTZ | 4 | CGTGGATTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GACTAATTAC-----TC-CCCC-GAAGA--AGTCGCTTAACAAA-G--------------GACTCT---GTTCTTGCAGGGATCGCATTCGCAC |
| droBia1 | scf7180000302261:3089215-3089316 + | LASTZ | 4 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTGATTTC-----------GAGTGGTTAT-----CG-CACG-GGGAA--AGCCACTAATGCAA-G--------------GGATCT---TGTGTTGCAGGGATCGGATTCGGAC |
| droTak1 | scf7180000415856:360180-360285 - | LASTZ | 4 | CGTGGATTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GAGTGGCTAT-----CCTCATT-AGGTA--GATGACTAACCCAA-G--------------GAATCTTGTCTTGTCGCAGGGATCGGATTCGCAC |
| droEle1 | scf7180000491026:48627-48723 + | LASTZ | 4 | CGTGGATTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GTTTGTTTTT-----CT-CAAC-CGAAT--G----GTAACCTAC----------------AATTTA---ACATTTCTAGGGATCGGATTCGCAC |
| droRho1 | scf7180000767553:111843-111943 + | LASTZ | 4 | CGTGGATTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GAACATCTTT-----CT-GGGG-GCAAT--CATAATTTATACAA----------------TATTTG---GGATTTCCAGGGACCGGATTCGCAC |
| droFic1 | scf7180000453904:1013475-1013577 - | LASTZ | 4 | CGTGGACTGGTTTGTGTGAGTACGG-----AT------CTGGCTTT-----------GGTTGC-GGT-T---AA-CCCT-GGGGAGTAGTGAGTAATGTGG----------------GCCTTC---CACTTTTCAGGGACAGGATTCGGAC |
| droKik1 | scf7180000301725:290060-290161 + | LASTZ | 5 | CGTGGACTGGTTTGTGTGAGTATGA-----AGGGGGT---TTTCTG-------------------GT-----AT-CCTTGAGGGG--GATATTTGACCTAA-AGGGAT----------TATGT---TTTCCTAAAGGGATCGAATTCGCAC |
| droAna3 | scaffold_12916:1707814-1707900 - | LASTZ | 3 | CGTGGACTGGTTTGTGTGAGTATGA-----GA------TGGATGGAGGGCTCAGTCCGTGTAG-------------------------TAACC-------------------------ACATC---CCCTTTCCAGAGACCGCATCCGCAC |
| droBip1 | scf7180000396728:369103-369188 - | LASTZ | 4 | CGTCGACTGGTTTGTGTGAGTATGG-----GA------CCCAGTCC-----------GTGTGT--------------------------------------AAACCACTGTT--ATAATCCAC---TTCCCTCCAGAGATCGCATCCGCAC |
| dp4 | chr4_group3:8989807-8989896 + | LASTZ | 5 | CGTCGACTGGTTTGTGTAAGTCCTG-----A-----------TCCC-----------GACTC-----------------------TC----CTCC----TTAGAGCTTCGCTTCATTGTGTCC---TTTGCTGCAGCGACCGCATTCGGAC |
| droPer1 | super_8:173764-173853 + | LASTZ | 5 | CGTCGACTGGTTTGTGTAAGTCCTG-----A-----------TCCC-----------GACTC-----------------------TC----CTCC----TTAGAGCTTCGCTTCATTGTGTCC---TTTGCTGCAGCGACCGCATCCGGAC |
| droWil1 | scaffold_180708:429041-429131 + | LASTZ | 5 | CGTGGATTGGTTGCTGTAAGCAATATAT--AT------CTCTTTTG-----------CATAAT-TGT---------ATA-ATGCC---------AACTGCT------------------AATT---TATTTTCCAGGGATCGGTTCCGCAC |
| droVir3 | scaffold_12723:5465392-5465486 - | LASTZ | 5 | TGTCGACTGGCTGCTGTATGTAAAT-----GC------TAGAACTT-----------TATGT-----------------------TT----GGTC----CTGGATATACATTTTCTGTTCGAT---TAATTTTTAGTGATCGTTTCCGCAC |
| droMoj3 | scaffold_6500:11048776-11048863 - | LASTZ | 5 | TGTCGATTGGCTACTGTATGTAGATGTC--AA---AATTTGTACTT-----------GACTGT-----------------------T----ATTTATTGAA-----------------ATTTG---ATTGTCTTAGGGATCGTTTCCGTAC |
| droGri2 | scaffold_15252:7120755-7120854 - | LASTZ | 5 | TGTCGATTGGTTTGTGTAAGTACTGGG-GGAGGGGAT-----TATG-----------GATTA-----------------------TC----CCAA----ATATAGTTTTGAATTTATAAATTC---TGATTTAAAGTGATCGCATTCGCAC |
| Species | Alignment |
| dm3 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTAATTTG-----------TAATGGCAGT-----TC-CACT-GCGAA--AAAGCTTTATCGAA-G--------------ATCCAT---CTCCTTGCAGGGATCGGATTCGCAC |
| droSim1 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GAATGGCAGT-----TC-CACT-GCGCC--AAAGCTTTATCGAA----------------GATTCT---TCTCTTGCAGGGATCGGATTCGCAC |
| droSec1 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GAATGGCAGT-----TC-CACT-GCACA--AAAGCTTTATCGAA----------------GATTCT---TCTCTTGCAGGGATCGGATTCGCAC |
| droYak2 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTGATTAG-----------GATTCC-AATGTAATTC-TACT-GCGAA--ACAGCTTCATCGAA----------------GGTTCT---TCTCTTGCAGGGATCGGATTCGCAC |
| droEre2 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CCGATTCA-----------GAATGCTGAT-----TC-CACT-GCGAA--GAAGCTTCATCGAA----------------GGGGCT---TCTCTTGCAGGGATCGGATTCGCAC |
| droEug1 | CGTGGATTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GACTAATTAC-----TC-CCCC-GAAGA--AGTCGCTTAACAAA-G--------------GACTCT---GTTCTTGCAGGGATCGCATTCGCAC |
| droBia1 | CGTGGACTGGTTTGTGTGAGTATGG-----AT------CTGATTTC-----------GAGTGGTTAT-----CG-CACG-GGGAA--AGCCACTAATGCAA-G--------------GGATCT---TGTGTTGCAGGGATCGGATTCGGAC |
| droTak1 | CGTGGATTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GAGTGGCTAT-----CCTCATT-AGGTA--GATGACTAACCCAA-G--------------GAATCTTGTCTTGTCGCAGGGATCGGATTCGCAC |
| droEle1 | CGTGGATTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GTTTGTTTTT-----CT-CAAC-CGAAT--G----GTAACCTAC----------------AATTTA---ACATTTCTAGGGATCGGATTCGCAC |
| droRho1 | CGTGGATTGGTTTGTGTGAGTATGG-----AT------CTGATTTG-----------GAACATCTTT-----CT-GGGG-GCAAT--CATAATTTATACAA----------------TATTTG---GGATTTCCAGGGACCGGATTCGCAC |
| droFic1 | CGTGGACTGGTTTGTGTGAGTACGG-----AT------CTGGCTTT-----------GGTTGC-GGT-T---AA-CCCT-GGGGAGTAGTGAGTAATGTGG----------------GCCTTC---CACTTTTCAGGGACAGGATTCGGAC |
| droKik1 | CGTGGACTGGTTTGTGTGAGTATGA-----AGGGGGT---TTTCTG-------------------GT-----AT-CCTTGAGGGG--GATATTTGACCTAA-AGGGAT----------TATGT---TTTCCTAAAGGGATCGAATTCGCAC |
| droAna3 | CGTGGACTGGTTTGTGTGAGTATGA-----GA------TGGATGGAGGGCTCAGTCCGTGTAG-------------------------TAACC-------------------------ACATC---CCCTTTCCAGAGACCGCATCCGCAC |
| droBip1 | CGTCGACTGGTTTGTGTGAGTATGG-----GA------CCCAGTCC-----------GTGTGT--------------------------------------AAACCACTGTT--ATAATCCAC---TTCCCTCCAGAGATCGCATCCGCAC |
| dp4 | CGTCGACTGGTTTGTGTAAGTCCTG-----A-----------TCCC-----------GACTC-----------------------TC----CTCC----TTAGAGCTTCGCTTCATTGTGTCC---TTTGCTGCAGCGACCGCATTCGGAC |
| droPer1 | CGTCGACTGGTTTGTGTAAGTCCTG-----A-----------TCCC-----------GACTC-----------------------TC----CTCC----TTAGAGCTTCGCTTCATTGTGTCC---TTTGCTGCAGCGACCGCATCCGGAC |
| droWil1 | CGTGGATTGGTTGCTGTAAGCAATATAT--AT------CTCTTTTG-----------CATAAT-TGT---------ATA-ATGCC---------AACTGCT------------------AATT---TATTTTCCAGGGATCGGTTCCGCAC |
| droVir3 | TGTCGACTGGCTGCTGTATGTAAAT-----GC------TAGAACTT-----------TATGT-----------------------TT----GGTC----CTGGATATACATTTTCTGTTCGAT---TAATTTTTAGTGATCGTTTCCGCAC |
| droMoj3 | TGTCGATTGGCTACTGTATGTAGATGTC--AA---AATTTGTACTT-----------GACTGT-----------------------T----ATTTATTGAA-----------------ATTTG---ATTGTCTTAGGGATCGTTTCCGTAC |
| droGri2 | TGTCGATTGGTTTGTGTAAGTACTGGG-GGAGGGGAT-----TATG-----------GATTA-----------------------TC----CCAA----ATATAGTTTTGAATTTATAAATTC---TGATTTAAAGTGATCGCATTCGCAC |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
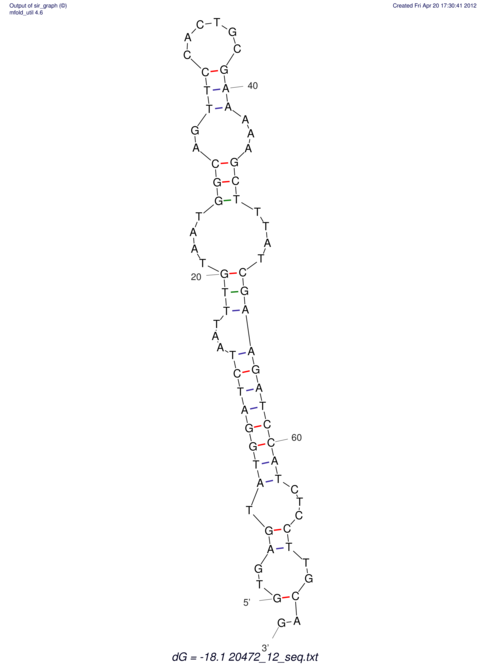
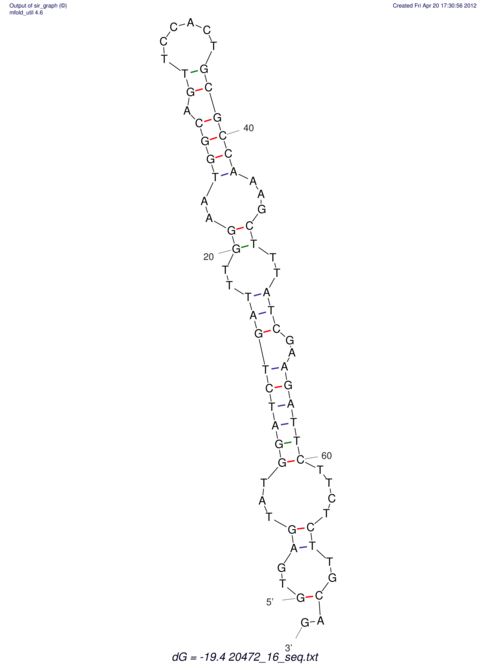
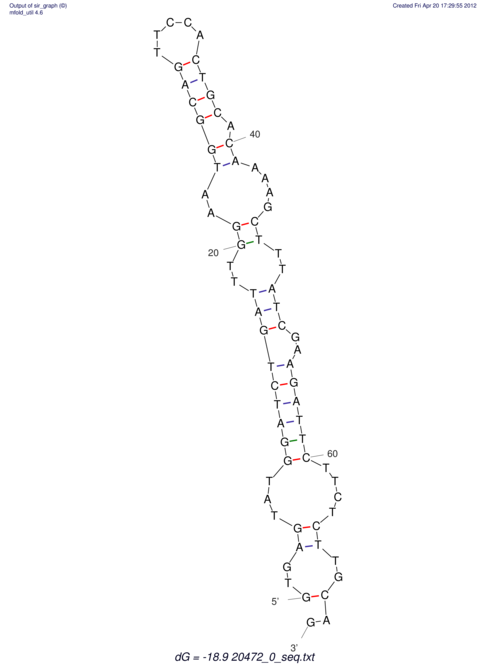
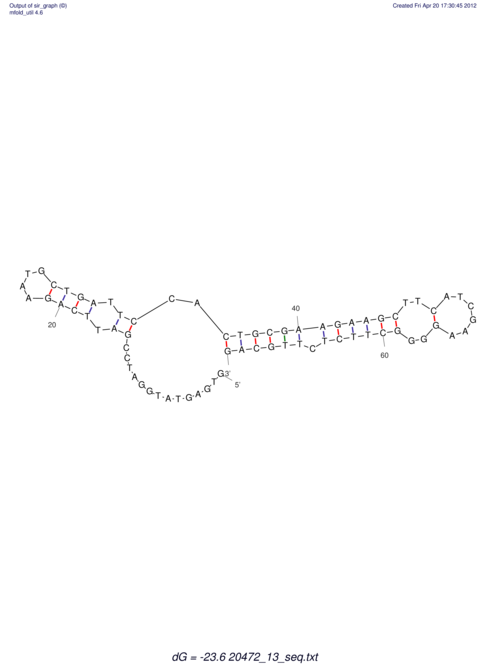
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
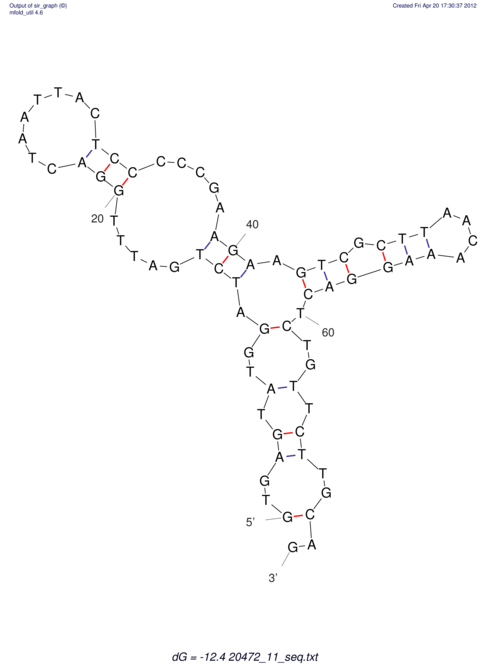
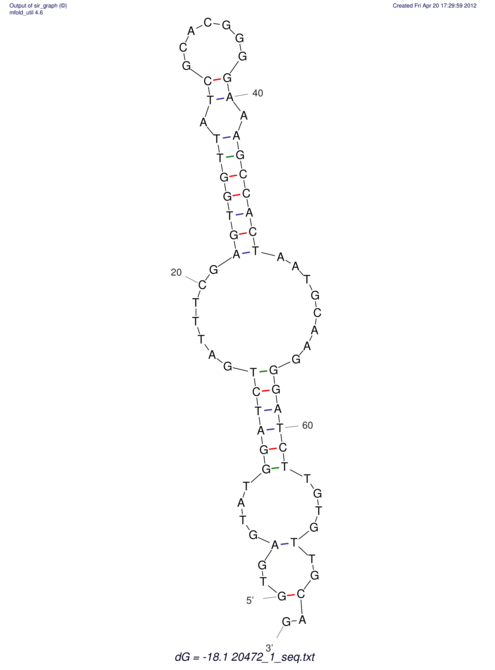
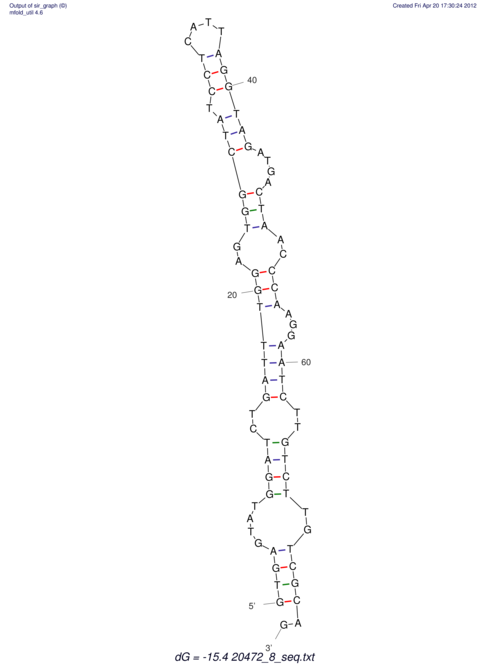
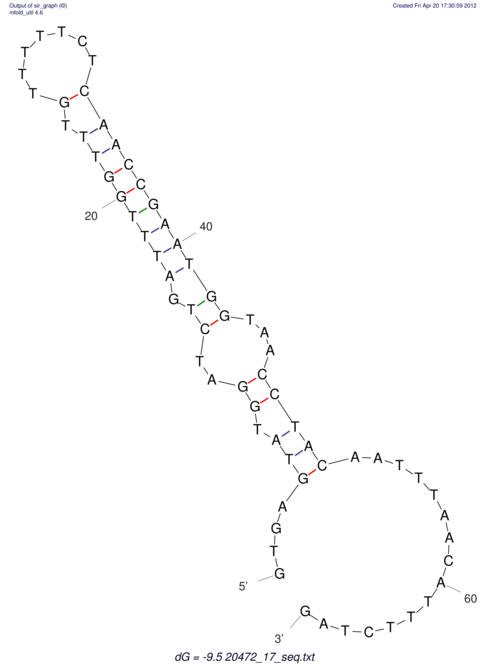
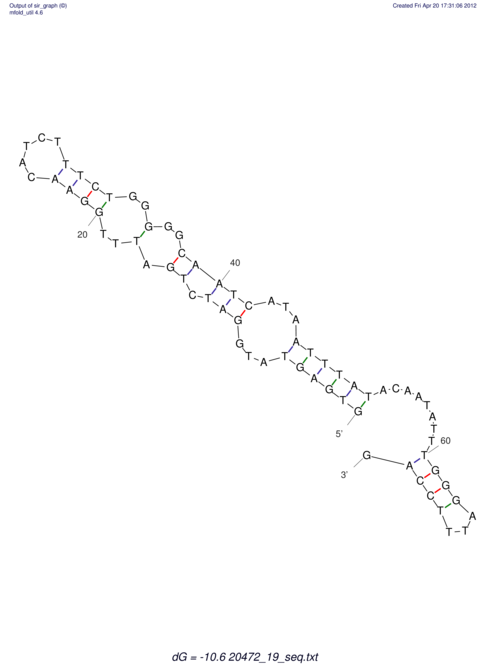
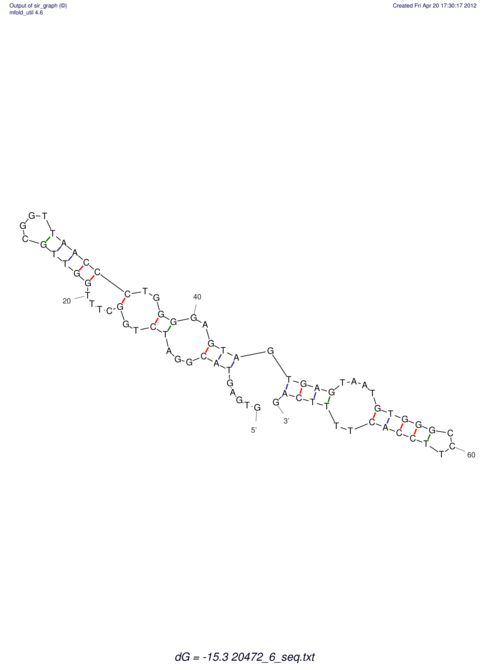
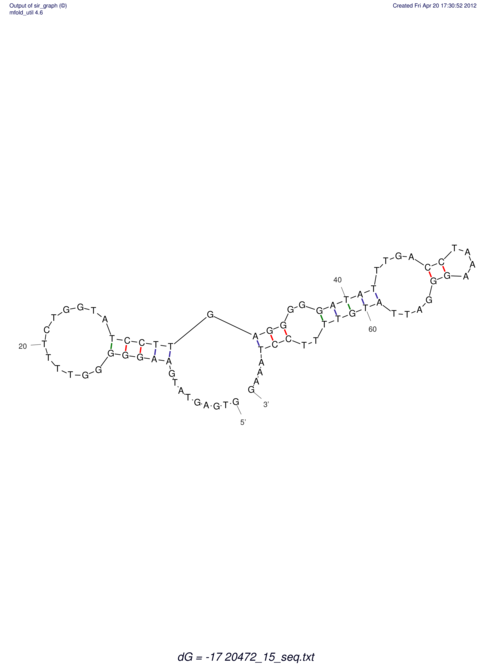
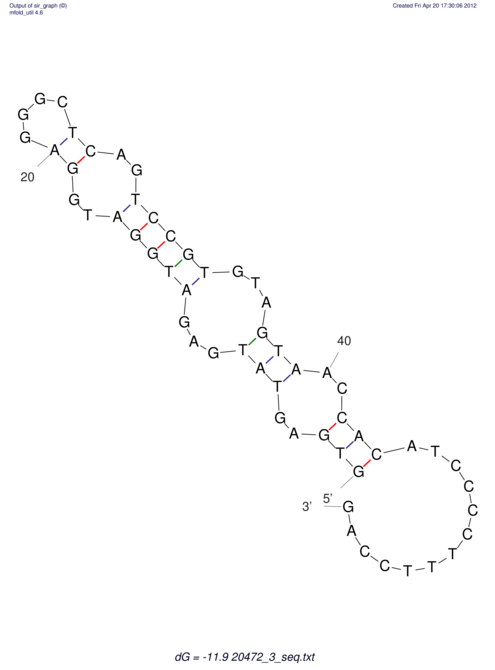
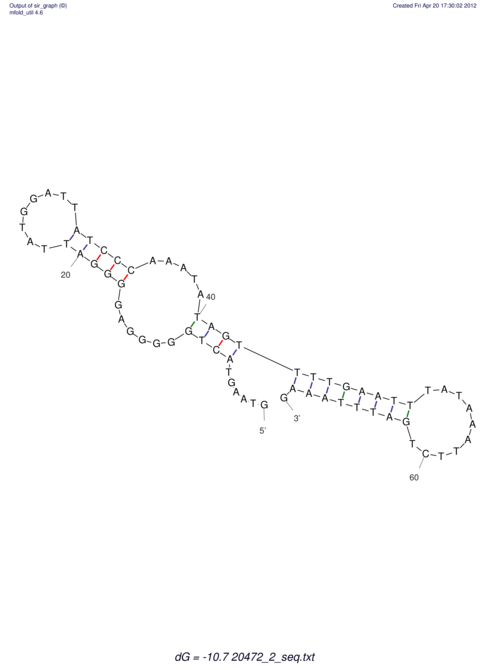
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:31:10 EDT