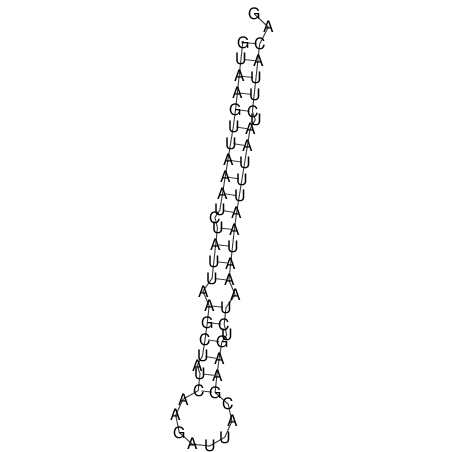
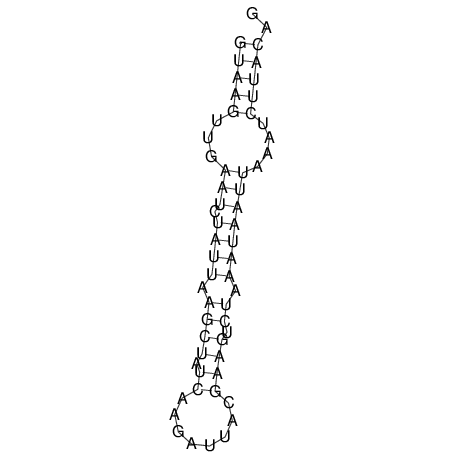
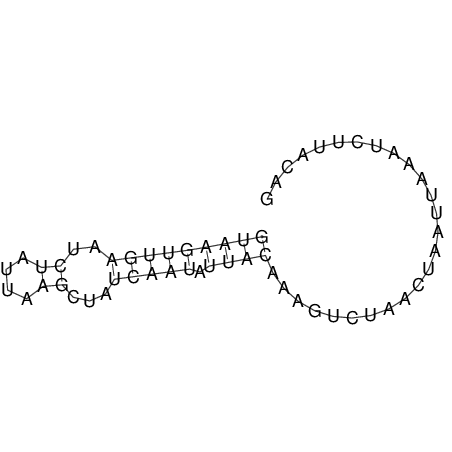
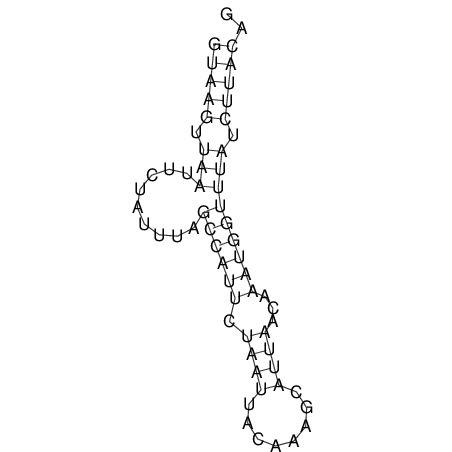
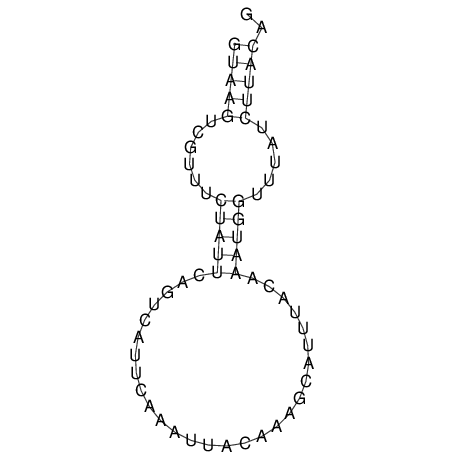
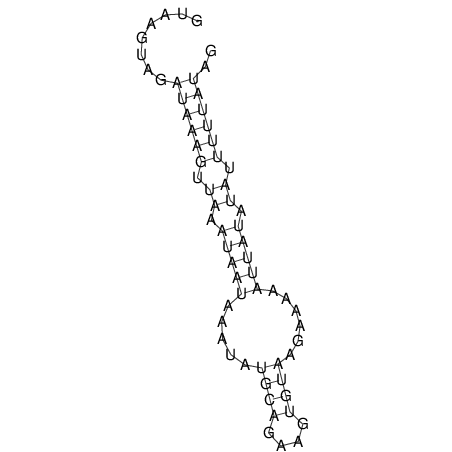
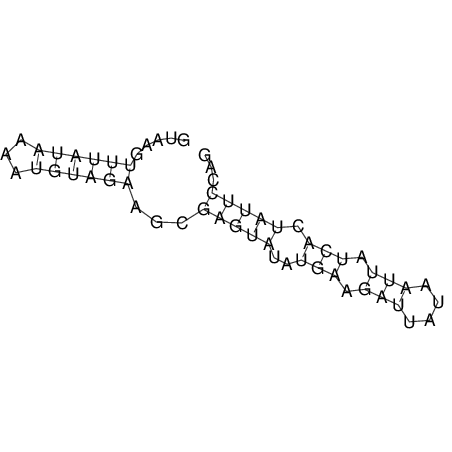
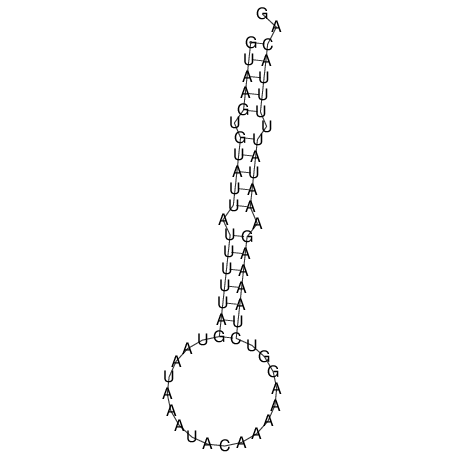
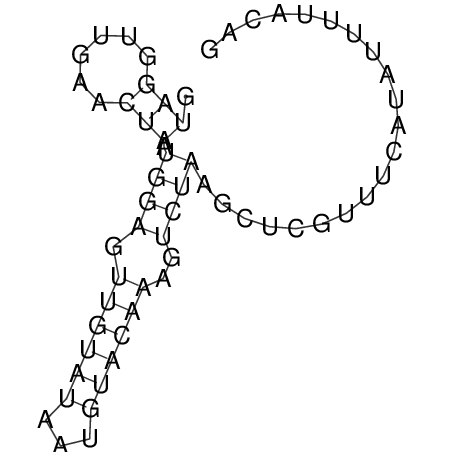
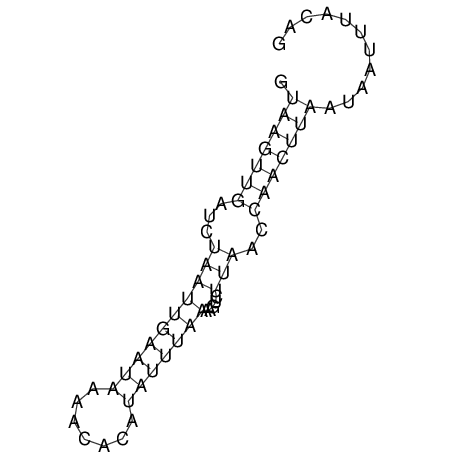
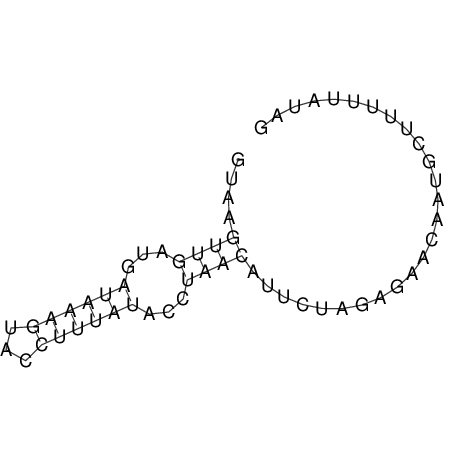
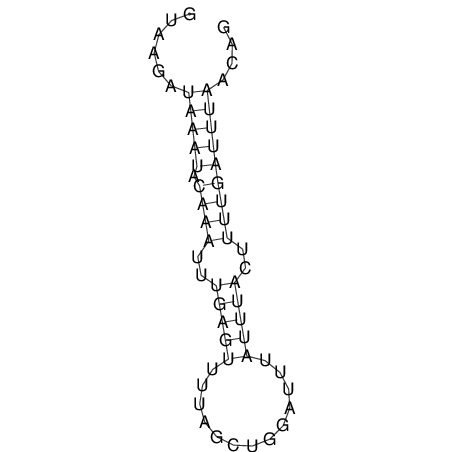
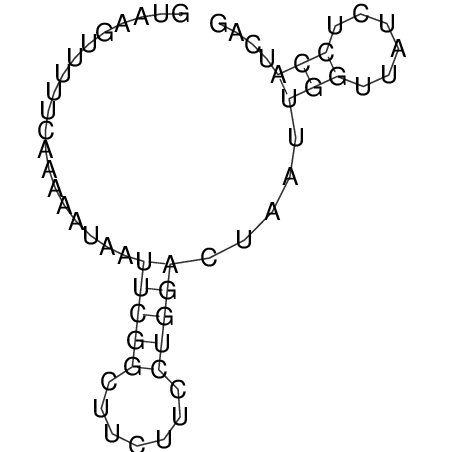
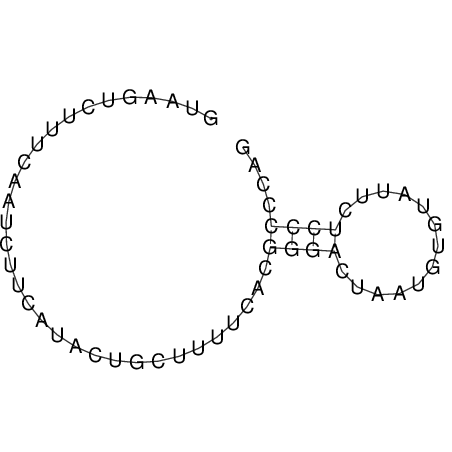
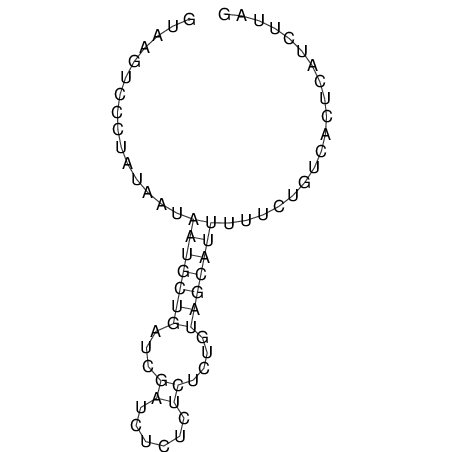
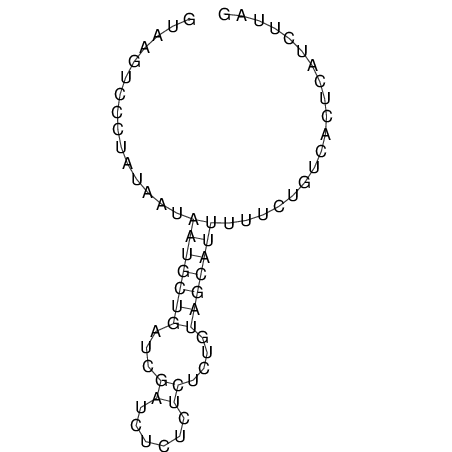

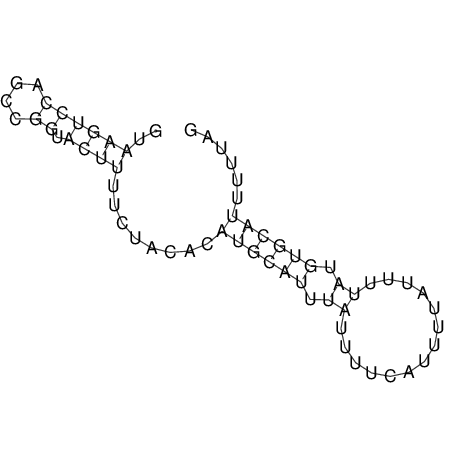
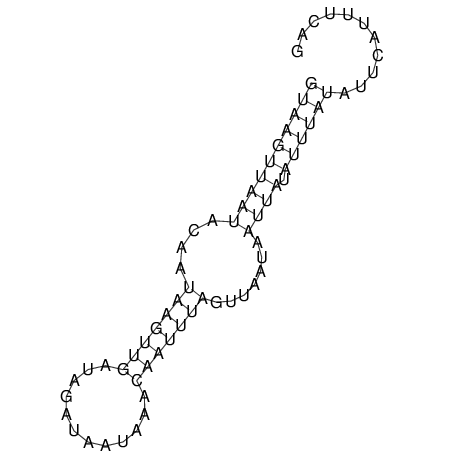
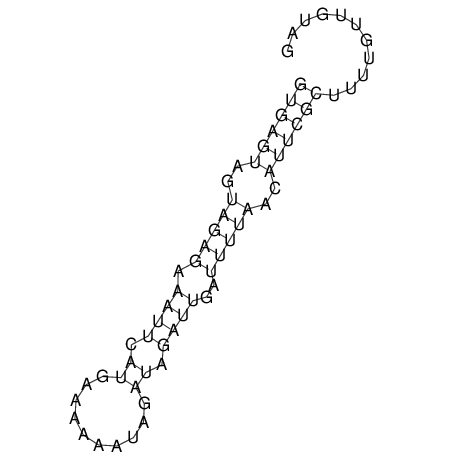
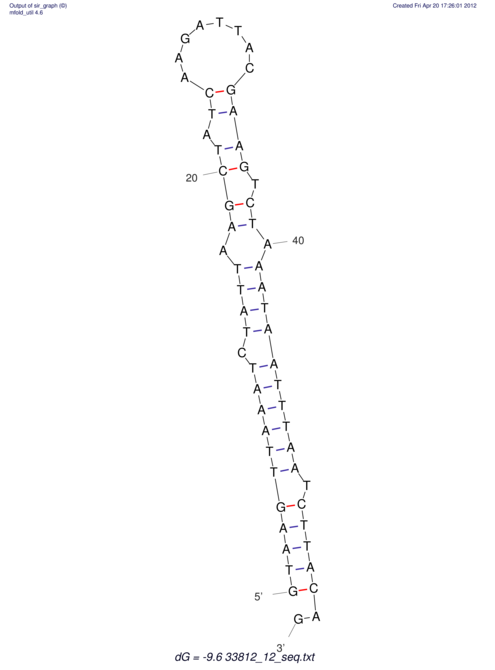
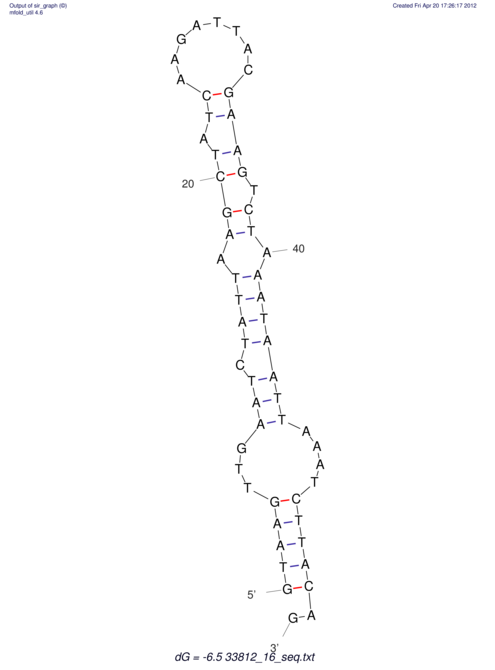


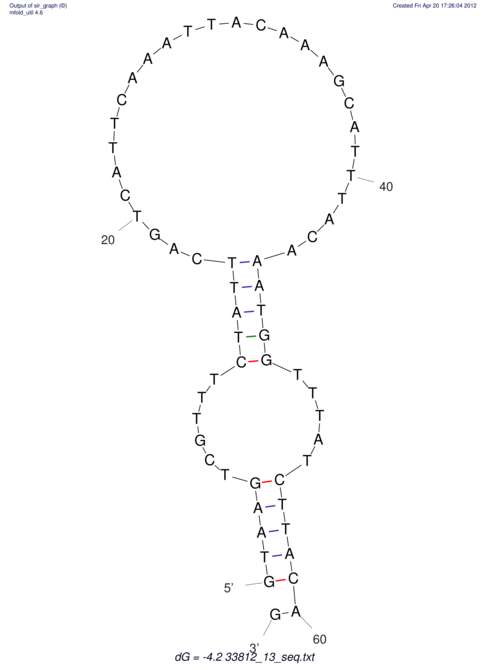
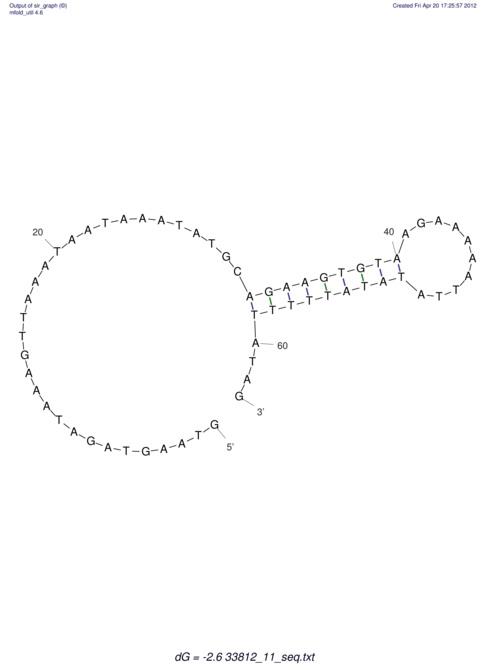
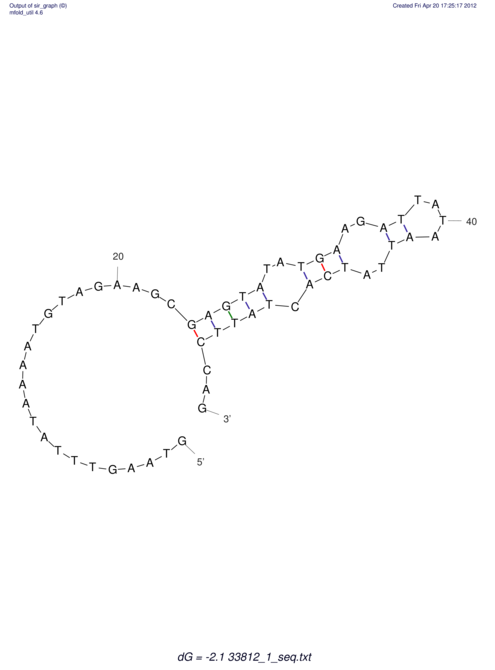
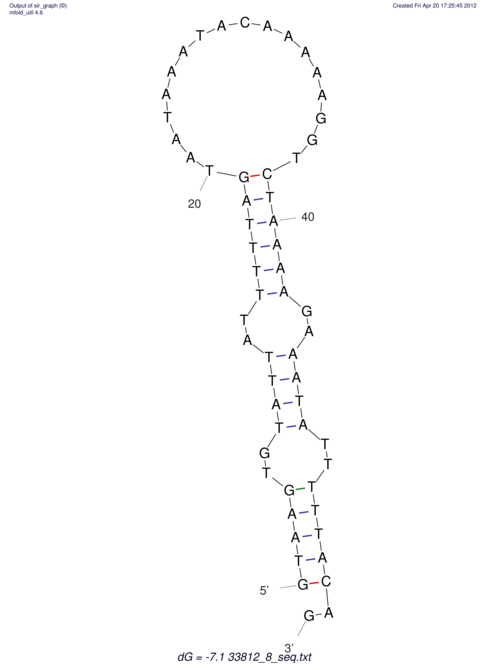
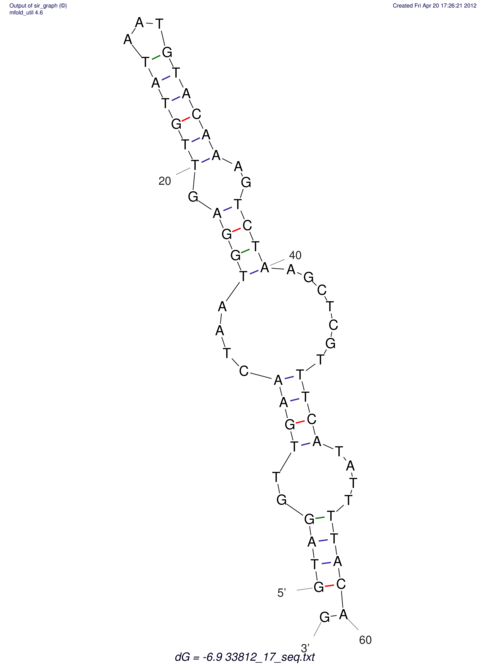
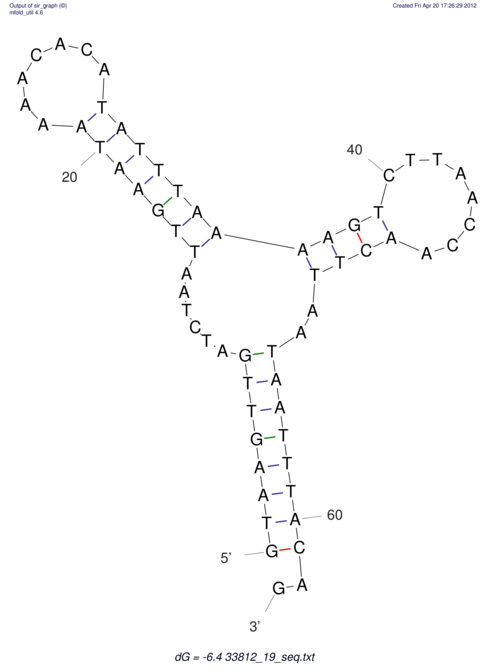
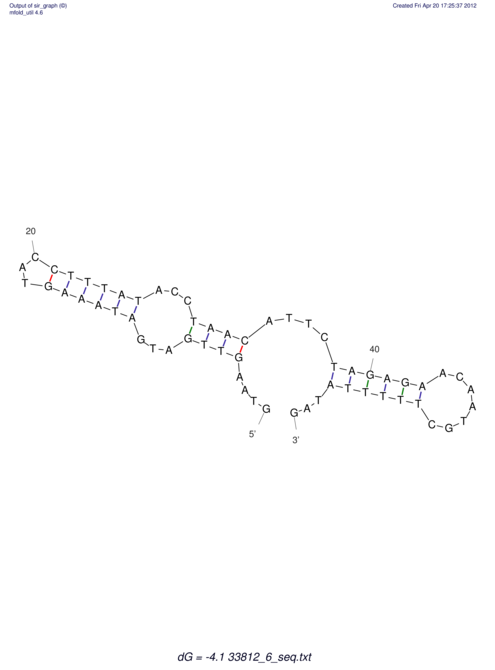


View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
CG15160_in3 |
chr2L:18592119-18592176 + |
Candidate |
|---|---|---|
| [View on UCSC Genome Browser {Cornell Mirror}] | mirtron |
Legend:
mature strand at position 18592119-18592139| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr2L:18592104-18592191 + | 1 | AAAGTGGTAGCCAGTGTAAGTTAAATCTAT----TAA-----GCTAT---CA--AGATT-ACGA------A-GT-----------CTAAATAA-----TTT--AATCTTACAGGCCTATAAGAGCTTT | |
| droSim1 | chr2L:18282956-18283043 + | LASTZ | 4 | AAAGTGGTAGCCAGTGTAAGTTGAATCTAT----TAA-----GCTAT---CA--AGATT-ACGA------A-GT-----------CTAAATAA-----TTA--AATCTTACAGGCCTATAAGAGCTTT |
| droSec1 | super_7:2217339-2217426 + | LASTZ | 5 | AAAGTGGTAGCCAGTGTAAGTTGAATCTAT----TAA-----GCTAT---CA--ATATT-ACAA------A-GT-----------CTAACTAA-----TTA--AATCTTACAGGCCTATAAGAGCTTT |
| droYak2 | chr2R:5109327-5109417 + | LASTZ | 5 | AAAGTGGTTGCCAGTGTAAGTTAATTCTAT----TTA-----GCCAT---TC--TAATT-ACAA------A-GC-----------ATTAACAAA-TGG-TT--TATCTTACAGGCCTATAAGAGCTTT |
| droEre2 | scaffold_4845:5679928-5680018 - | LASTZ | 5 | AAAGTGGTCGCCAGCGTAAGTCGTTTCTAT----TCA-----GTCAT---TC--AAATT-ACAA------A-GC-----------ATTTACAAA-TGG-TT--TATCTTACAGGCCTATAAGAGCTTT |
| droEug1 | scf7180000409447:540216-540308 + | LASTZ | 5 | AAGGTGGTTGCCAGTGTAAGTAGATAAAGT----TAA-----ATAAT---AA--ATATG-CAGA------A-GT-----------GTAAGAAAAATTATAT--ATTTTTATAGGCCTATAAAAGTTTT |
| droBia1 | scf7180000302422:5087976-5088062 - | LASTZ | 5 | AAGGTGGTGGCCAGCGTAAGTTTATAAAAT----GTA-----GAAGC-GAG-----TAT-ATGA------A-GA-----------TTATAATT-----ATC--ACTATTCCAGGCCTATAAAAGTTTC |
| droTak1 | scf7180000415867:892356-892443 + | LASTZ | 4 | AAGGTGGTGGCCAGTGTAAGTGTATTATTT----TTA-----GTAAT---AA--ATACA-AAAA------G-GT-----------CTAAAAGA-----AAT--ATTTTTACAGGCCTATAAAAGTTTT |
| droEle1 | scf7180000491338:2569757-2569847 + | LASTZ | 4 | AAAGTGGTGGCCAGTGTAGGTTGAACTAAT----GGA-----GTTGT---AT--AATGT-ACAA------A-GT-----------CTAAGCTCG-TTT-CA--TATTTTACAGGCCTATAAAAGCTTT |
| droRho1 | scf7180000778026:24955-25047 + | LASTZ | 5 | AAAGTGGTGGCCAGTGTAAGTTGATCTAAT----TGA-----ATAAA---ACACATATT-TAAA------A-GT-----------CTTAACCAA-CTT-AA--TAATTTACAGGCCTATAAAAGCTTT |
| droFic1 | scf7180000454059:343979-344067 - | LASTZ | 5 | AAAGTGGTGGCCAGTGTAAGTTGATGATAAAGTA-CC-------------T-----TTAT-AC-CTAACA--TTCTAGAG------------AA-CAA-TG--CTTTTTATAGGCCTATAAAAGTTTT |
| droKik1 | scf7180000302221:23696-23781 - | LASTZ | 4 | AAGGTGGTGGCCAGTGTAAGATAAATACAA------------ATT--TGAG-----TTTT-A--------------GCTGG----ATTTATTTA-C---TTTTGATTTAACAGGCCTACAAGAGCTTC |
| droAna3 | scaffold_12916:2495263-2495349 + | LASTZ | 5 | AAAGTGGTGGCAAGTGTAAGTTTTTCAAAA----ATA-----ATTCG--GC-----TTCTTCCT------G-GA-----------CTAATTGG-----TTA--TCTCCATCAGGCCTATAAGAGCTTC |
| droBip1 | scf7180000396430:434558-434644 + | LASTZ | 5 | AAGGTGGTGGCAAGTGTAAGTCTTTCAATC----TTC-----ATACT--GC-----TTTTCACG------G-GA-----------CTAATGTG-----TAT--TCTCCCCCAGGCCTACAAAAGTTTC |
| dp4 | chr4_group3:3803651-3803745 + | LASTZ | 5 | AAAGTGGTGGCCAGTGTAAGTCCCTATAATAATG-CTGATCGATC--TCTC-----TCTC-T--------------GTAGC----ATTTTTCTG-T---CA--CTCATCTTAGGCCTATAAAAGTTTC |
| droPer1 | super_1:5283458-5283552 + | LASTZ | 5 | AAAGTGGTGGCCAGTGTAAGTCCCTATAATAATG-CTGATCGATC--TCTC-----TCTC-T--------------GTAGC----ATTTTTCTG-T---CA--CTCATCTTAGGCCTATAAAAGTTTC |
| droWil1 | scaffold_180772:3053226-3053306 - | LASTZ | 5 | AG--TCCTTAAATG------CCATATCTAA----TTC-----TATAT---CG--TTATA-TCTA------A-AC----------TTCAAATTT-----CTC--TTTATCAAAGGCCTACAAAAGTTTT |
| droVir3 | scaffold_12963:18639372-18639466 + | LASTZ | 5 | AAAGTTGTCGCAAGTGTAAGTCCAGCCGGT----ACT-----TTTCT---AC--ACATG-CATT------TATT----------TTCATTTTATTTTATGT--GCATTTTTAGGCCTACAAAAGCTTT |
| droMoj3 | scaffold_6500:6015082-6015180 - | LASTZ | 5 | AAAGTTGTGGCAAGCGTAAGTTAATACAATAA-G-TTGATAGATA--ATAA-----AC------------A-ATTTAGTTAATAATTATATTT-----ATA--TTCATTTCAGGCTTATAAAAGTTTT |
| droGri2 | scaffold_15252:6570131-6570224 + | LASTZ | 4 | AAAGTGGTTGCCAGTGTGAGTAGTAGAGAA-A-TTCA-----TGAAA---AAATAGATA-GATT------GATT----------TTAACATTCG-----CT--TTTGTTGTAGGCCTATAAAAGTTTT |
| Species | Alignment |
| dm3 | AAAGTGGTAGCCAGTGTAAGTTAAATCTAT----TAA-----GCTAT---CA--AGATT-ACGA------A-GT-----------CTAAATAA-----TTT--AATCTTACAGGCCTATAAGAGCTTT |
| droSim1 | AAAGTGGTAGCCAGTGTAAGTTGAATCTAT----TAA-----GCTAT---CA--AGATT-ACGA------A-GT-----------CTAAATAA-----TTA--AATCTTACAGGCCTATAAGAGCTTT |
| droSec1 | AAAGTGGTAGCCAGTGTAAGTTGAATCTAT----TAA-----GCTAT---CA--ATATT-ACAA------A-GT-----------CTAACTAA-----TTA--AATCTTACAGGCCTATAAGAGCTTT |
| droYak2 | AAAGTGGTTGCCAGTGTAAGTTAATTCTAT----TTA-----GCCAT---TC--TAATT-ACAA------A-GC-----------ATTAACAAA-TGG-TT--TATCTTACAGGCCTATAAGAGCTTT |
| droEre2 | AAAGTGGTCGCCAGCGTAAGTCGTTTCTAT----TCA-----GTCAT---TC--AAATT-ACAA------A-GC-----------ATTTACAAA-TGG-TT--TATCTTACAGGCCTATAAGAGCTTT |
| droEug1 | AAGGTGGTTGCCAGTGTAAGTAGATAAAGT----TAA-----ATAAT---AA--ATATG-CAGA------A-GT-----------GTAAGAAAAATTATAT--ATTTTTATAGGCCTATAAAAGTTTT |
| droBia1 | AAGGTGGTGGCCAGCGTAAGTTTATAAAAT----GTA-----GAAGC-GAG-----TAT-ATGA------A-GA-----------TTATAATT-----ATC--ACTATTCCAGGCCTATAAAAGTTTC |
| droTak1 | AAGGTGGTGGCCAGTGTAAGTGTATTATTT----TTA-----GTAAT---AA--ATACA-AAAA------G-GT-----------CTAAAAGA-----AAT--ATTTTTACAGGCCTATAAAAGTTTT |
| droEle1 | AAAGTGGTGGCCAGTGTAGGTTGAACTAAT----GGA-----GTTGT---AT--AATGT-ACAA------A-GT-----------CTAAGCTCG-TTT-CA--TATTTTACAGGCCTATAAAAGCTTT |
| droRho1 | AAAGTGGTGGCCAGTGTAAGTTGATCTAAT----TGA-----ATAAA---ACACATATT-TAAA------A-GT-----------CTTAACCAA-CTT-AA--TAATTTACAGGCCTATAAAAGCTTT |
| droFic1 | AAAGTGGTGGCCAGTGTAAGTTGATGATAAAGTA-CC-------------T-----TTAT-AC-CTAACA--TTCTAGAG------------AA-CAA-TG--CTTTTTATAGGCCTATAAAAGTTTT |
| droKik1 | AAGGTGGTGGCCAGTGTAAGATAAATACAA------------ATT--TGAG-----TTTT-A--------------GCTGG----ATTTATTTA-C---TTTTGATTTAACAGGCCTACAAGAGCTTC |
| droAna3 | AAAGTGGTGGCAAGTGTAAGTTTTTCAAAA----ATA-----ATTCG--GC-----TTCTTCCT------G-GA-----------CTAATTGG-----TTA--TCTCCATCAGGCCTATAAGAGCTTC |
| droBip1 | AAGGTGGTGGCAAGTGTAAGTCTTTCAATC----TTC-----ATACT--GC-----TTTTCACG------G-GA-----------CTAATGTG-----TAT--TCTCCCCCAGGCCTACAAAAGTTTC |
| dp4 | AAAGTGGTGGCCAGTGTAAGTCCCTATAATAATG-CTGATCGATC--TCTC-----TCTC-T--------------GTAGC----ATTTTTCTG-T---CA--CTCATCTTAGGCCTATAAAAGTTTC |
| droPer1 | AAAGTGGTGGCCAGTGTAAGTCCCTATAATAATG-CTGATCGATC--TCTC-----TCTC-T--------------GTAGC----ATTTTTCTG-T---CA--CTCATCTTAGGCCTATAAAAGTTTC |
| droWil1 | AG--TCCTTAAATG------CCATATCTAA----TTC-----TATAT---CG--TTATA-TCTA------A-AC----------TTCAAATTT-----CTC--TTTATCAAAGGCCTACAAAAGTTTT |
| droVir3 | AAAGTTGTCGCAAGTGTAAGTCCAGCCGGT----ACT-----TTTCT---AC--ACATG-CATT------TATT----------TTCATTTTATTTTATGT--GCATTTTTAGGCCTACAAAAGCTTT |
| droMoj3 | AAAGTTGTGGCAAGCGTAAGTTAATACAATAA-G-TTGATAGATA--ATAA-----AC------------A-ATTTAGTTAATAATTATATTT-----ATA--TTCATTTCAGGCTTATAAAAGTTTT |
| droGri2 | AAAGTGGTTGCCAGTGTGAGTAGTAGAGAA-A-TTCA-----TGAAA---AAATAGATA-GATT------GATT----------TTAACATTCG-----CT--TTTGTTGTAGGCCTATAAAAGTTTT |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
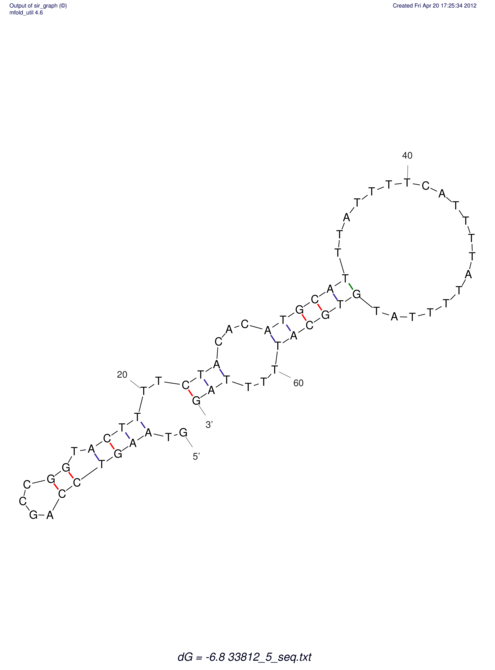
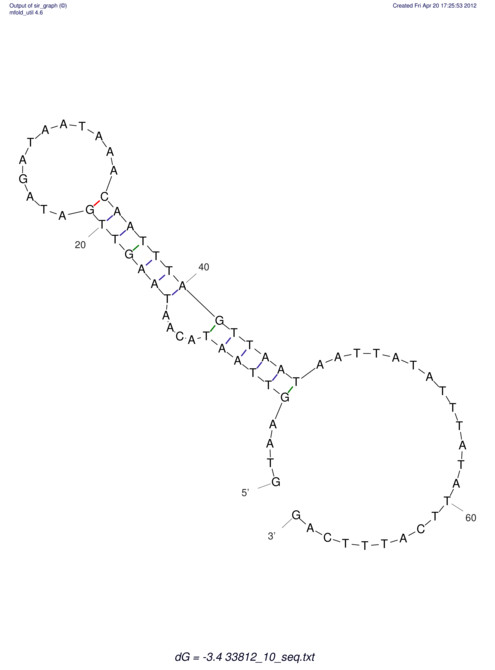
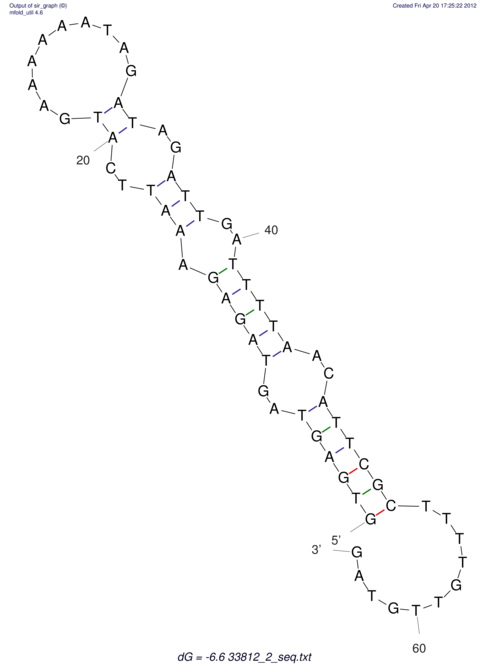
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||
| droSim1 |
|
||||||||||||||||||
| droSec1 |
|
||||||||||||||||||
| droYak2 |
|
||||||||||||||||||
| droEre2 |
|
||||||||||||||||||
| droEug1 |
|
||||||||||||||||||
| droBia1 |
|
||||||||||||||||||
| droTak1 |
|
||||||||||||||||||
| droEle1 |
|
||||||||||||||||||
| droRho1 |
|
||||||||||||||||||
| droFic1 |
|
||||||||||||||||||
| droKik1 |
|
||||||||||||||||||
| droAna3 |
|
||||||||||||||||||
| droBip1 |
|
||||||||||||||||||
| dp4 |
|
||||||||||||||||||
| droPer1 |
|
||||||||||||||||||
| droWil1 |
|
||||||||||||||||||
| droVir3 |
|
||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:26:32 EDT