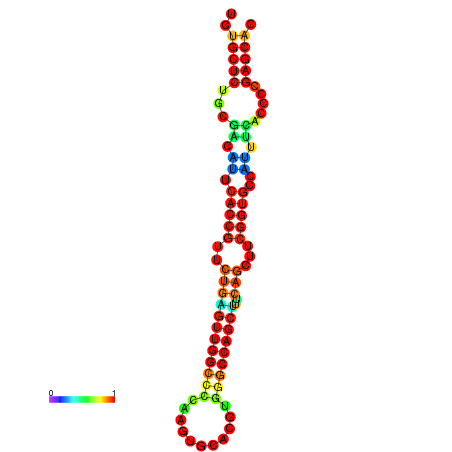
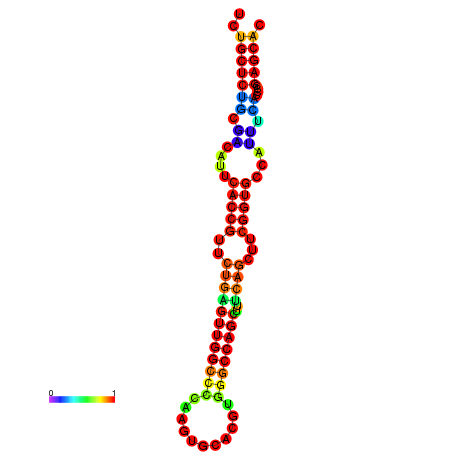
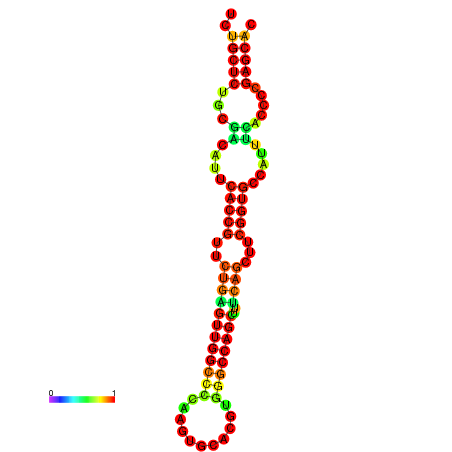
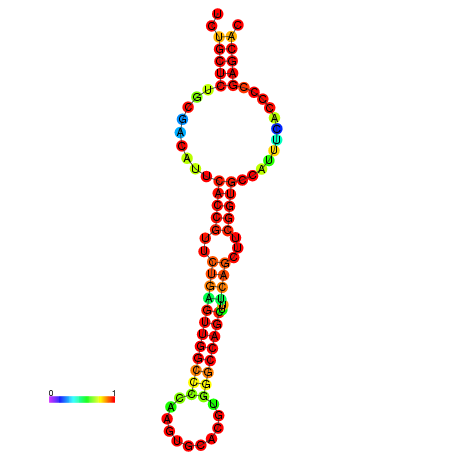
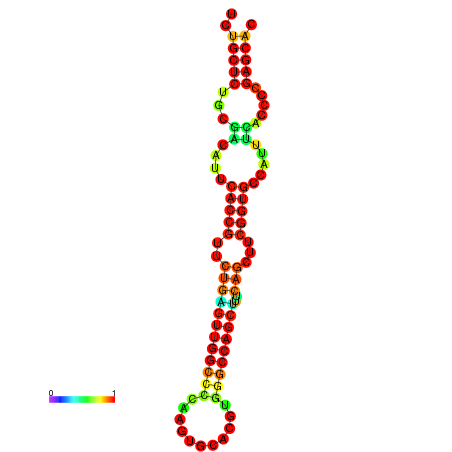
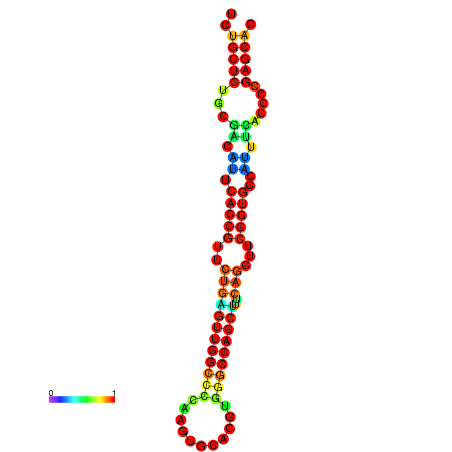
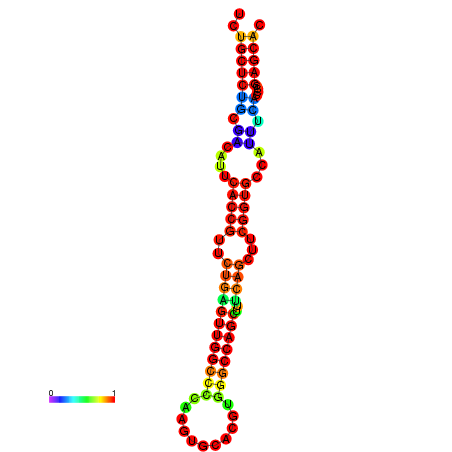
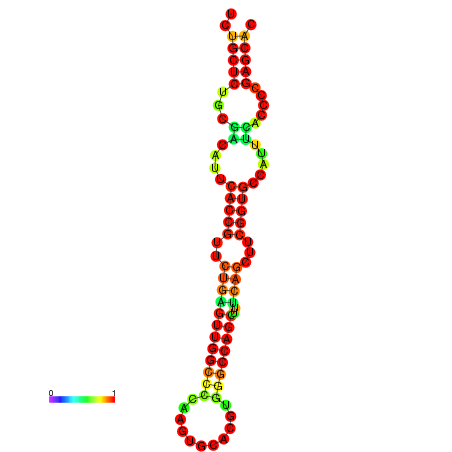
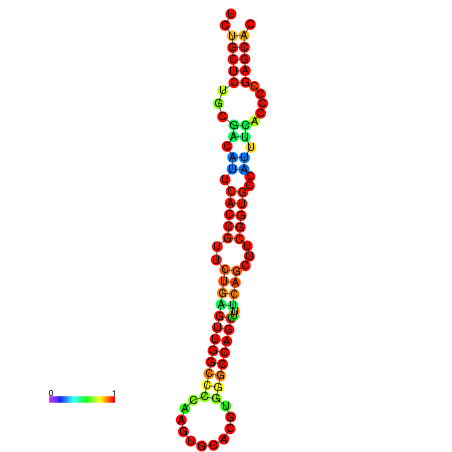
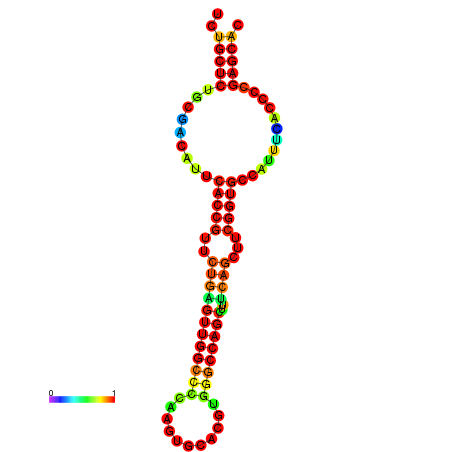
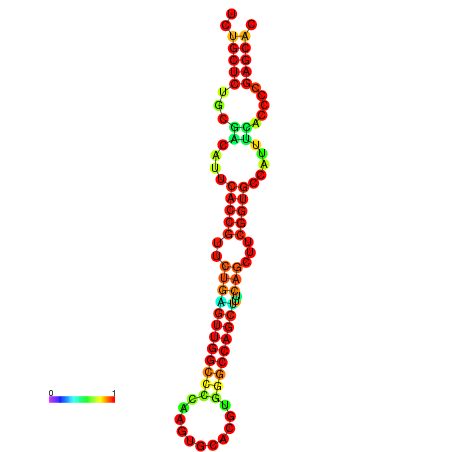
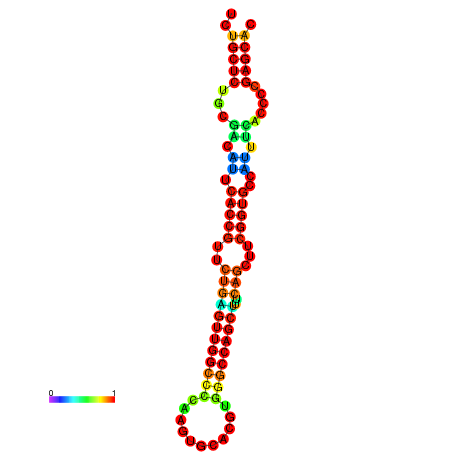
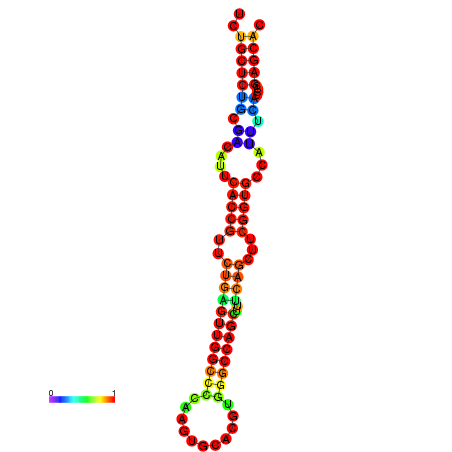
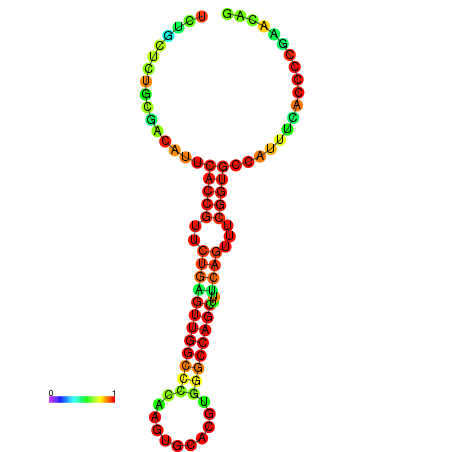
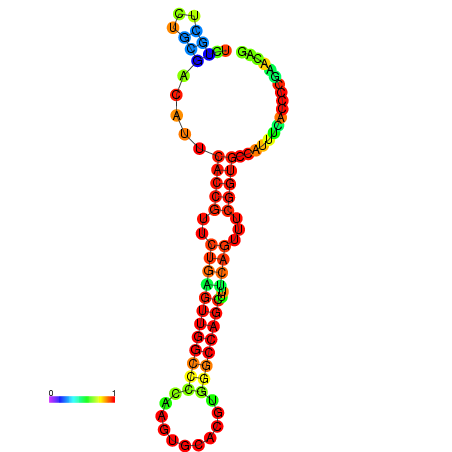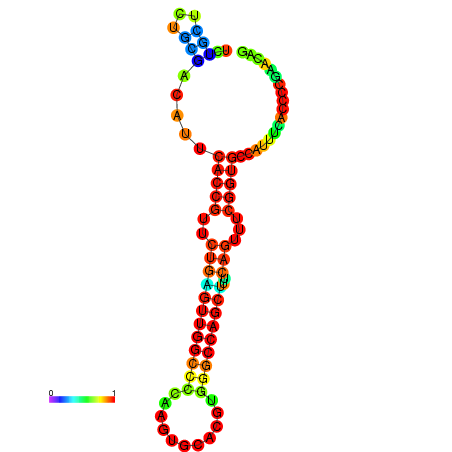| dm3 |
chr2R:12782713-12782828 - |
TGGGAAAC-----------TGGCGA----ATCTG------------CTCTGC-GACATTCACCGTTCTGAGTTGGCCCCAAGTGCACGTGGGCCAG--------------CTTTCAGCTTCGGTGCC-ATTT-----------------------------------------CACCC------------CGAGCACCAAGAGCATCAGACT |
| droSim1 |
chr2R:11518341-11518456 - |
TGGGAAAC-----------TGGCGA----ATCTG------------CTCTGC-GACATTCACCGTTCTGAGTTGGCCCCAAGTGCACGTGGGCCAG--------------CTTTCAGCTTCGGTGCC-ATTT-----------------------------------------CACCC------------CGAGCACCAAGAGCATCAGACT |
| droSec1 |
super_1:10286785-10286900 - |
TGGGAAAC-----------TGGCGA----ATCTG------------CTCTGC-GACATTCACCGTTCTGAGTTGGCCCCAAGTGCACGTGGGCCAG--------------CTTTCAGCTTCGGTGCC-ATTT-----------------------------------------CACCC------------CGAGCACCAAGAGTATCAGACT |
| droYak2 |
chr2R:16191830-16191945 + |
TGGGAAAC-----------TGGCGA----ATCTG------------CTCTGC-GACATTCACCGTTCTGAGTTGGCCCCAAGTGCACGTGGGCCAG--------------CTTTCAGTTTCGGTGCC-ATTT-----------------------------------------CACCC------------CGAACAGCAAGACCATCAGACT |
| droEre2 |
scaffold_4845:13064860-13064974 + |
TGGGTAAC-----------TGGCGA----GTCTG------------CTCTGC-GACATTCACCGTTCTGAGTTGGCCCCAAGTGCACGTGGGCCAG--------------CTTTCAGTTTCGGTGCC-ATTT-----------------------------------------CACCC------------CCAGCACCAAGAGCACC-GACT |
| droAna3 |
scaffold_13266:13815719-13815830 + |
TGGGAAAT-----------TGTCGA----AGCCA------------CTTTGC-GACATTCCCCGTTCTGCGTTGGCCCAAGGTGCACGTGGGGCCGACGA----------CTCTCCGCTTCCGGACCCATTG--------------------------------------------------------------CCGAAAATACATTCTTCA |
| dp4 |
chr3:6727705-6727891 - |
TGGGAAAC-----------TGGCGA----GGCG-AGGCGAGGACTCCTCTGC-GACATTCACCGTTCAGCGTTCGGCCAGCGAGCACGTGGGTTAC--GCCTGAAGCTCCAATTCGGTTACGGT------TTCGGCTACGGCTACGGCTACGGGTACGGCTACGGCTACGGTACATCCTCCACACTAGACGGCGGGTTGAGGGGGTCGGACA |
| droPer1 |
super_2:6950668-6950854 - |
TGGGAAAC-----------TGGCGA----GGCG-AGGCGAGGACTCCTCTGC-GACATTCACCGTTCAGCGTTCGGCCAGCGAGCACGTGGGTTAC--GCCTGAAGCTCCAATTCGGTTACGGT------TTCGGCTACGGCTACGGCTACGGGTACGGCTACGGCTACGGTACATCCTCCACACTAGACGGCGGGTTGAGGGGGTCGGACA |
| droWil1 |
scaffold_180700:6478759-6478837 - |
TGGGAAATACATCTTCGTTTGGCTTTAGTTTCTC------------TTCTGCGGACATTCAATGTTCTGCGTTTGCCCCAAGCGCACGTGG------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_12875:2573949-2574010 + |
TGGGAAAC-----------TG-CGA----AGCTG------------CTTTGC-GACATTCGACTTGCTGCGTTCGTCGCCGACGCACGTGG------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6496:22657688-22657749 - |
TGGGAAAC------------GGCGA----AGCTT------------CTGTGC-GACATTCAACGTTCTGCGTTCGTCGCCGACGCACGTGG------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_15112:4702153-4702224 + |
TGGGAATT-----------TTGCGA----AGCTGCTGCTGTGGCT--GCTGC-GACATTCAACGTTCTGCGTTGGCCGCCGACGCACGTG-------------------------------------------------------------------------------------------------------------------------- |