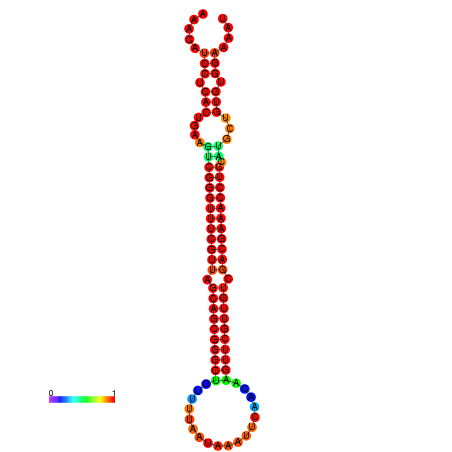
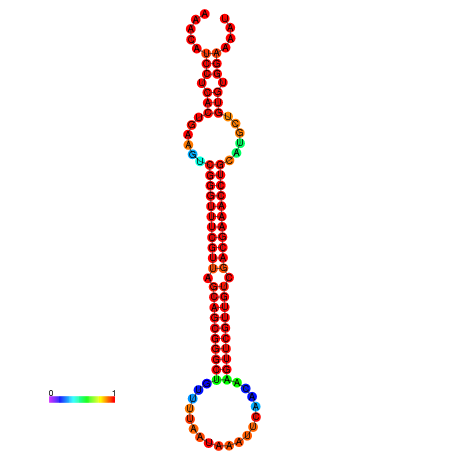
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:1682879-1683005 - | A-CTGCAACTTAGGAG-------------------AAACATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAATAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCAAGGCCG |
| droSim1 | chrX_random:1085378-1085505 - | AACTGCAACTTAGGAT-------------------AAACATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAATAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCAAGGCCA |
| droSec1 | super_10:1477754-1477881 - | AACTCCAACTTAGGAT-------------------AAACATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAATAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCAAGGCCA |
| droYak2 | chrX:1528063-1528198 - | A------ACTTAAGAGAGAGACCAA-----GTGCAAAGCATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAAAAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCAAGGCCG |
| droEre2 | scaffold_4644:1660954-1661093 - | A-CTGCAACTTAAGAGGG-CACCAA-----CTGCAAAGCATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAATAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCTAGGCCG |
| droAna3 | scaffold_12929:1586917-1587049 + | A-TTATAGTTTTA-----AGA---ACTGTGTTTCATAAAGTCCTCACTGAAGCCGGGTTTCGTTAACAGCGGGCTGTCATTTCGAA--------ATCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAATAACAACGTTTAA----- |
| dp4 | chrXL_group1a:4258914-4259036 - | A-CGAATATCTA-------------------TGCGAAAAATCCACATTGAAATCGGGTTTCGTTAACAGCGGGCTGTTATGAATAA--------ATCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAGAAACAAC----AAATGCA |
| droPer1 | super_11:835055-835162 - | T---------G-------------------ATGCGAAAAATCCACATTGAAATCGGGTTTCGTTAACAGCGGGCTGTTATGAATAA--------ATCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAGAAACAAC----------- |
| droWil1 | scaffold_181096:4683233-4683341 + | ---------------------------------------ATCCACACTGAAATCGGGTTTCGCTAGCAGCGGACTGTCATGATAAATGTGAAAAACAAACAAGTTCGTTGTCGACGAAACCTGCATCATGTGTCGGACAAACAGCTCT-------- |
| droVir3 | scaffold_12942:51501-51599 + | A-----------------------------------ACAATCTACATTGAACTCAGATTTCGTCAGCAGCGGGCTGTGATGATTA----------TGAACAAGTTCGTTGTCGACGAAACCTGCATACTGTGT-GGAGTGGCAAC----------- |
| droMoj3 | scaffold_6328:4397474-4397588 + | --CTGTGGATAAAAAT-------------------AGTAATCTACATTGAACTCAGGTTTCGTTAGCAACGGGCTGTGATGATTAA--------AAGAACAAGTTCGTTGTCGACGAAACCTGCATGCTATGT-GGAGAGGCAAC----------- |
| droGri2 | scaffold_14853:795296-795391 + | ----------------------------------------TCCACATTGAGCTCGGGTTTCGTCAGCAGCGGGCTGTGATGACTAA--------A-AAACAAGTTCGTTGTCGACGAAACCTGCATACTGTGT-GGAGAGTCAACG---------- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:1682879-1683005 - | A-CTGCAACTTAGGAG-------------------AAACATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAATAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCAAGGCCG |
| droSim1 | chrX_random:1085378-1085505 - | AACTGCAACTTAGGAT-------------------AAACATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAATAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCAAGGCCA |
| droSec1 | super_10:1477754-1477881 - | AACTCCAACTTAGGAT-------------------AAACATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAATAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCAAGGCCA |
| droYak2 | chrX:1528063-1528198 - | A------ACTTAAGAGAGAGACCAA-----GTGCAAAGCATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAAAAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCAAGGCCG |
| droEre2 | scaffold_4644:1660954-1661093 - | A-CTGCAACTTAAGAGGG-CACCAA-----CTGCAAAGCATCCTCACTGAAGTCGGGTTTCGTTAGCAGCGGGCTGTTTTAATAAA--------TTCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAAAATCAACGCTCTAGGCCG |
| droAna3 | scaffold_12929:1586917-1587049 + | A-TTATAGTTTTA-----AGA---ACTGTGTTTCATAAAGTCCTCACTGAAGCCGGGTTTCGTTAACAGCGGGCTGTCATTTCGAA--------ATCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAATAACAACGTTTAA----- |
| dp4 | chrXL_group1a:4258914-4259036 - | A-CGAATATCTA-------------------TGCGAAAAATCCACATTGAAATCGGGTTTCGTTAACAGCGGGCTGTTATGAATAA--------ATCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAGAAACAAC----AAATGCA |
| droPer1 | super_11:835055-835162 - | T---------G-------------------ATGCGAAAAATCCACATTGAAATCGGGTTTCGTTAACAGCGGGCTGTTATGAATAA--------ATCAACAAGTTCGTTGTCGACGAAACCTGCATGCTGTGT-GGAGAAACAAC----------- |
| droWil1 | scaffold_181096:4683233-4683341 + | ---------------------------------------ATCCACACTGAAATCGGGTTTCGCTAGCAGCGGACTGTCATGATAAATGTGAAAAACAAACAAGTTCGTTGTCGACGAAACCTGCATCATGTGTCGGACAAACAGCTCT-------- |
| droVir3 | scaffold_12942:51501-51599 + | A-----------------------------------ACAATCTACATTGAACTCAGATTTCGTCAGCAGCGGGCTGTGATGATTA----------TGAACAAGTTCGTTGTCGACGAAACCTGCATACTGTGT-GGAGTGGCAAC----------- |
| droMoj3 | scaffold_6328:4397474-4397588 + | --CTGTGGATAAAAAT-------------------AGTAATCTACATTGAACTCAGGTTTCGTTAGCAACGGGCTGTGATGATTAA--------AAGAACAAGTTCGTTGTCGACGAAACCTGCATGCTATGT-GGAGAGGCAAC----------- |
| droGri2 | scaffold_14853:795296-795391 + | ----------------------------------------TCCACATTGAGCTCGGGTTTCGTCAGCAGCGGGCTGTGATGACTAA--------A-AAACAAGTTCGTTGTCGACGAAACCTGCATACTGTGT-GGAGAGTCAACG---------- |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| -37.6 | -37.6 | -37.3 | -37.3 | -37.1 | -36.8 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
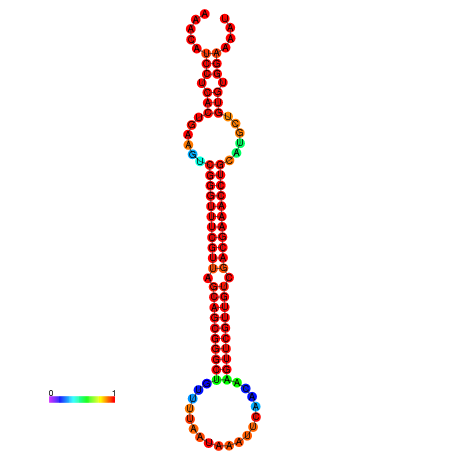 |
| -37.6 | -37.6 | -37.3 | -37.3 | -37.1 | -36.8 |
|---|---|---|---|---|---|
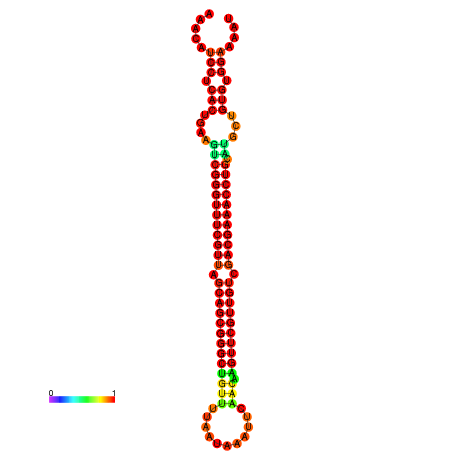 |
 |
 |
 |
 |
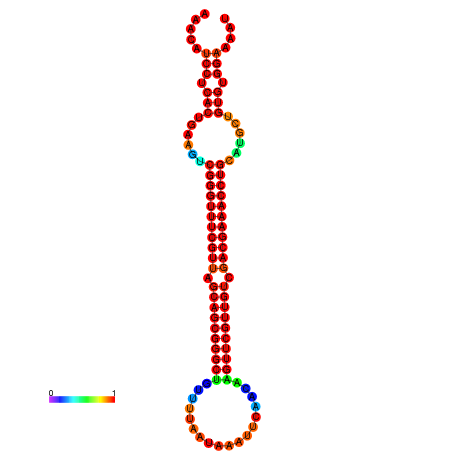 |
| -37.6 | -37.6 | -37.3 | -37.3 | -37.1 | -36.8 |
|---|---|---|---|---|---|
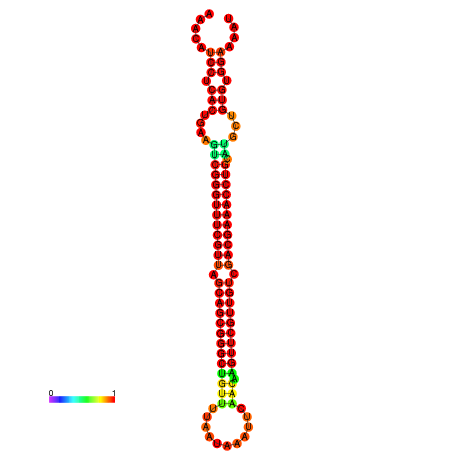 |
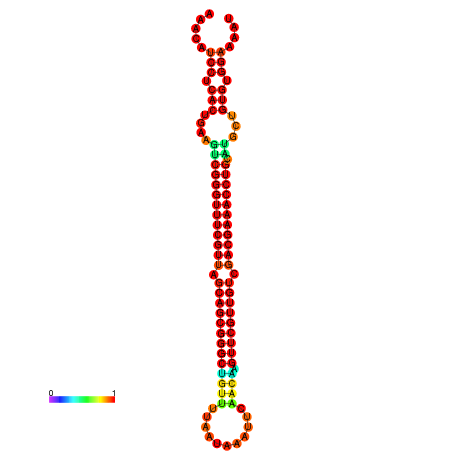 |
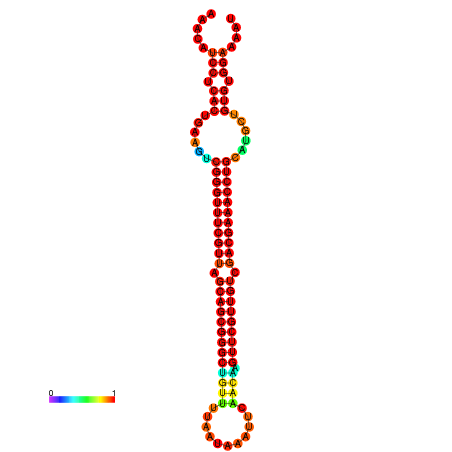 |
 |
 |
 |
| -37.6 | -37.6 | -37.3 | -37.3 | -37.1 | -36.8 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
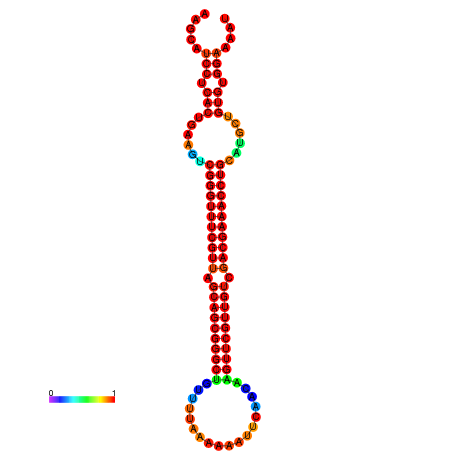 |
| -37.6 | -37.6 | -37.3 | -37.3 | -37.1 | -36.8 |
|---|---|---|---|---|---|
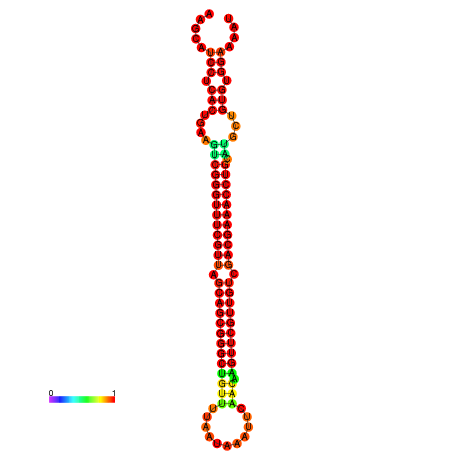 |
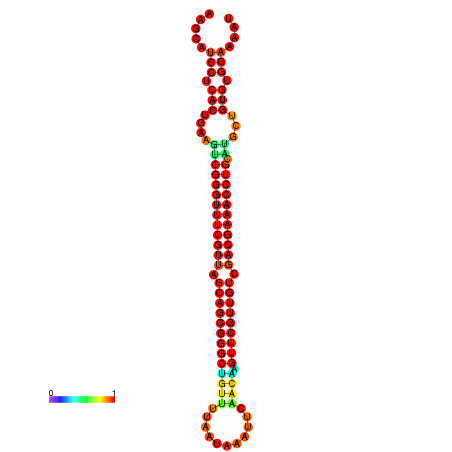 |
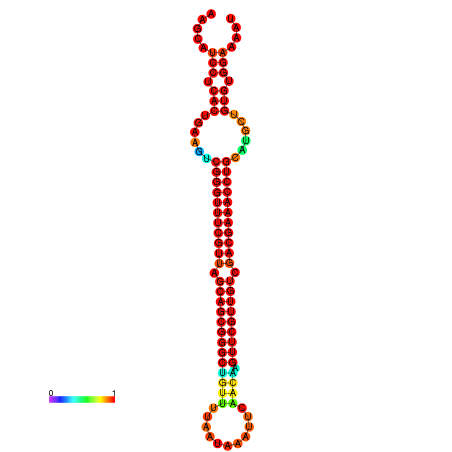 |
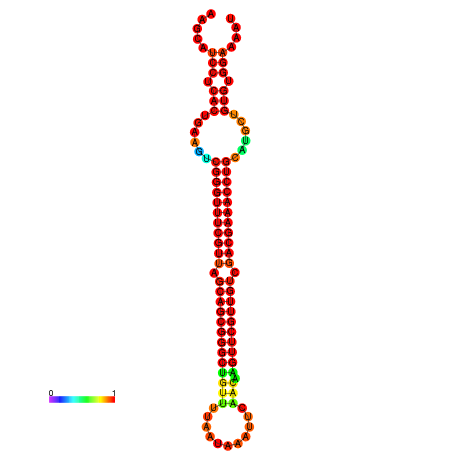 |
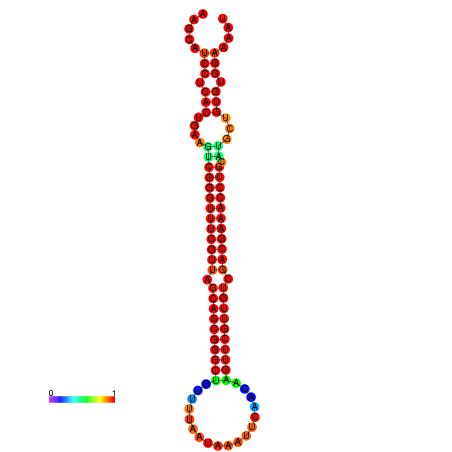 |
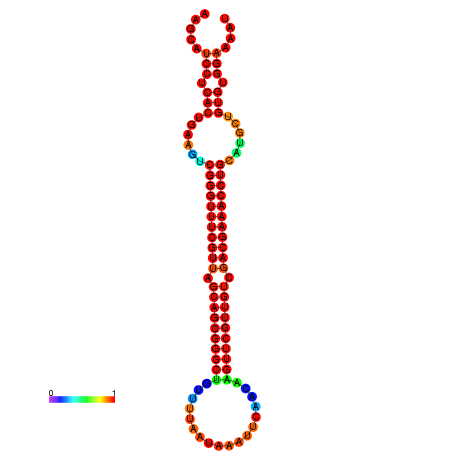 |
| -37.3 | -36.8 | -36.8 | -36.6 | -36.3 | -36.3 |
|---|---|---|---|---|---|
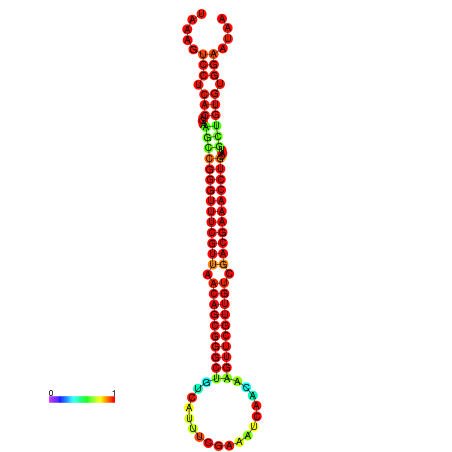 |
 |
 |
 |
 |
 |
| -41.0 | -41.0 | -41.0 | -41.0 | -40.9 | -40.9 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
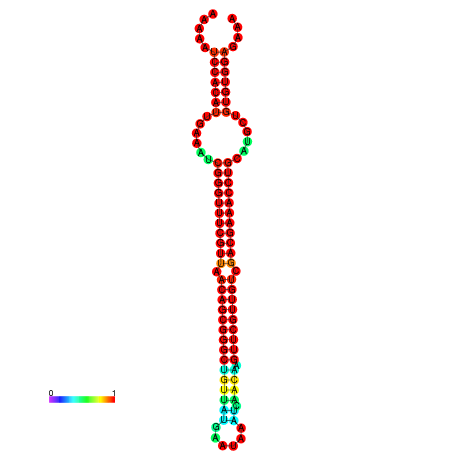
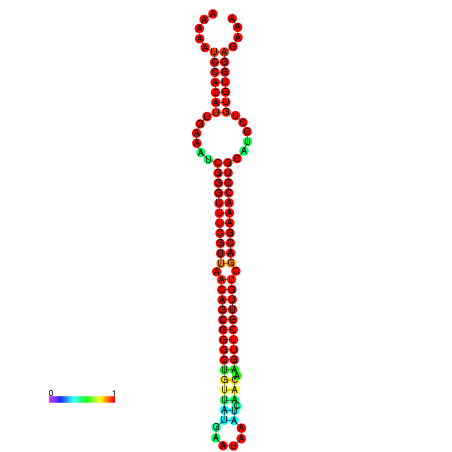
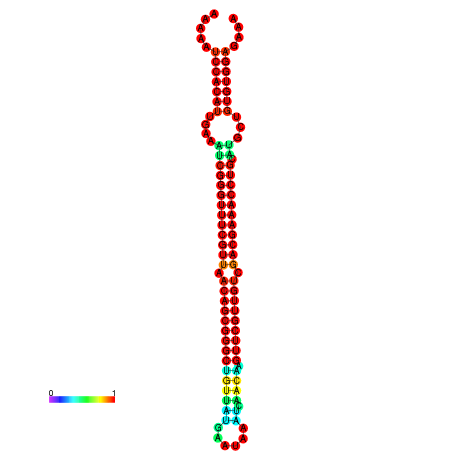
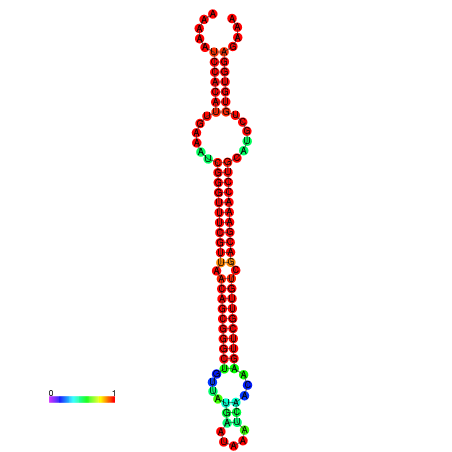
| -41.0 | -41.0 | -41.0 | -41.0 | -40.9 | -40.9 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
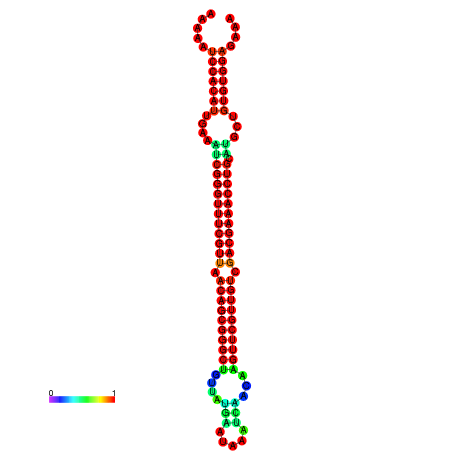 |
| -34.5 | -34.5 | -34.4 | -34.4 | -34.2 | -34.1 |
|---|---|---|---|---|---|
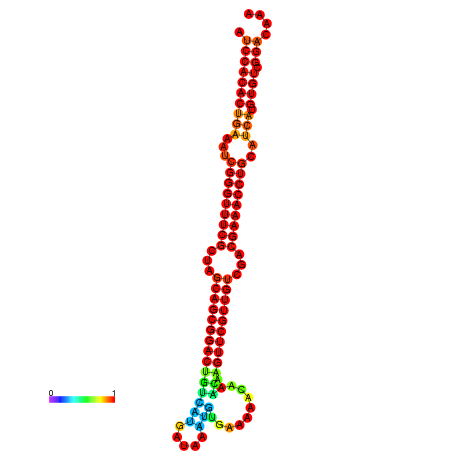 |
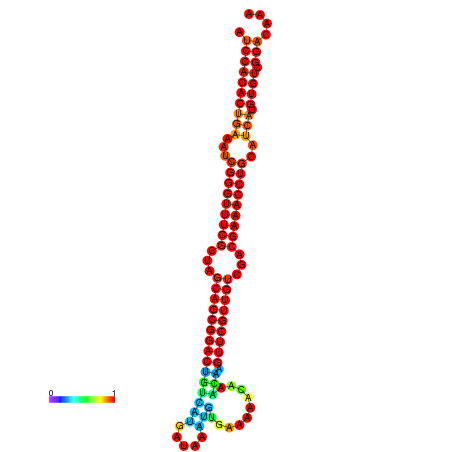 |
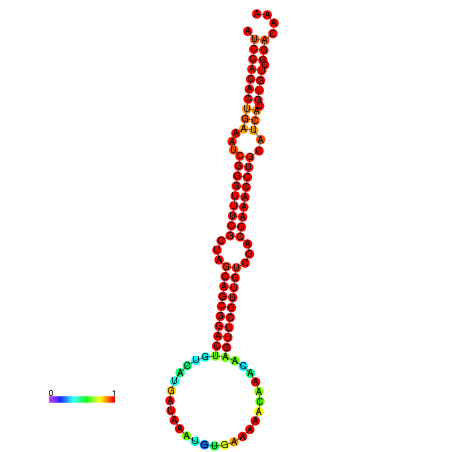 |
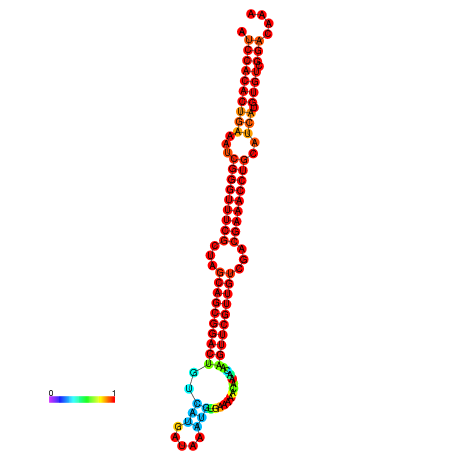 |
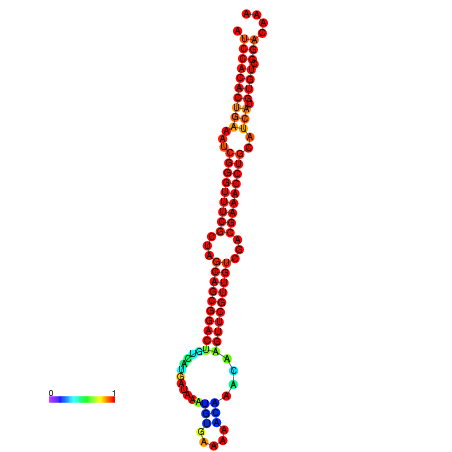 |
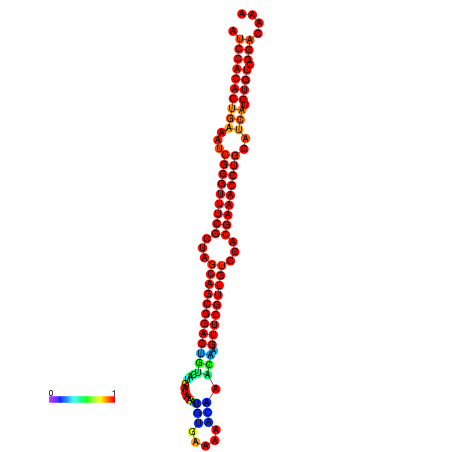 |
| -35.0 | -35.0 | -34.1 | -34.0 | -34.0 |
|---|---|---|---|---|
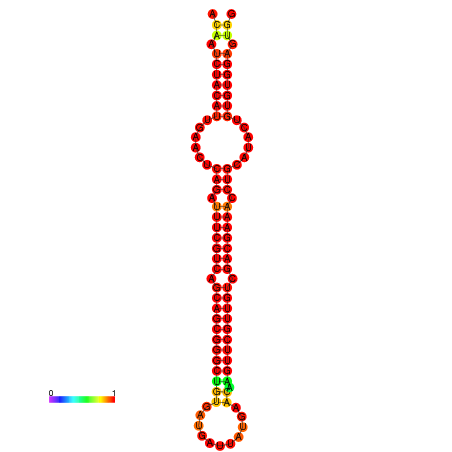 |
 |
 |
 |
 |
| -38.5 | -38.5 | -37.7 |
|---|---|---|
 |
 |
 |
| -41.8 | -41.8 | -40.9 |
|---|---|---|
 |
 |
 |
Generated: 01/05/2013 at 05:35 PM