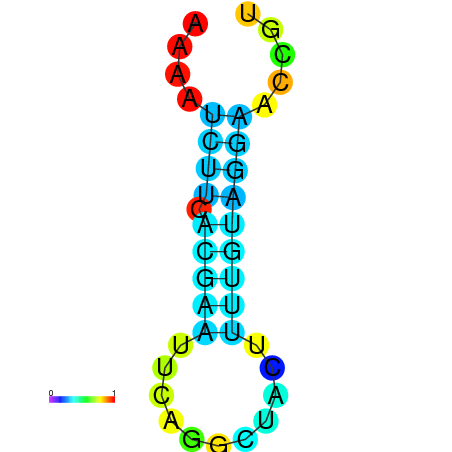
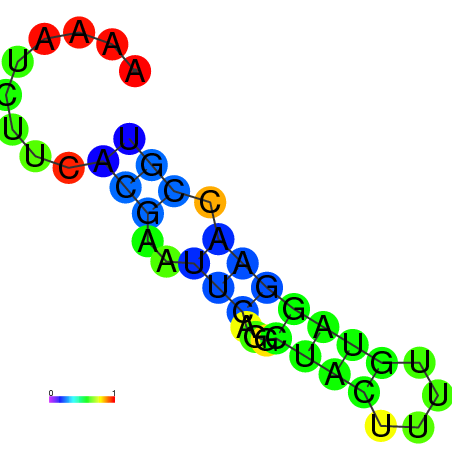
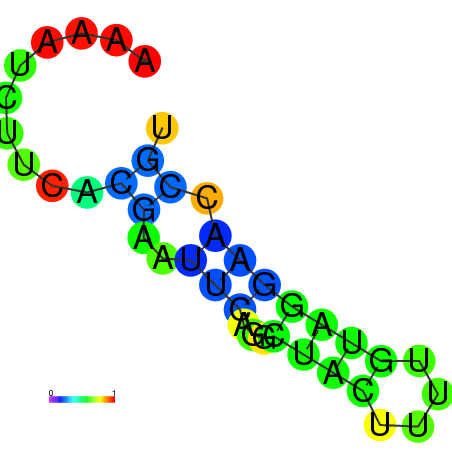
| dm3 |
chrX:19455855-19455979 + |
TTAATCAC---TGGAA-----------------------CATACATGTGAG-----------GATGTCACAAATACA--------------CTGA---ATTTGGGGGGAATTCT--------TATGTATATAC-AAATTCTTCCCGAACTCAGGCTAATTTTGTG---------------GCA---------------------------TCCGTATACACAAAA----------------------------------GT------ATA |
| droSim1 |
chrX_random:5079602-5079664 + |
TTAATATA---C--------------------------------------------------------------------------------------------------------------------------AAATTCTTCACGAATTCAGGCTAATTTTGTA---------------GGA---------------------------GCCGTATATACAAAA----------------------------------GT------ATA |
| droSec1 |
super_8:1738459-1738521 + |
TTAATATA---C--------------------------------------------------------------------------------------------------------------------------AAAATCTTCACGAATTCAGGCTACTTTTGTA---------------GGA---------------------------ACCGTATATACAAAA----------------------------------GT------ATA |
| droYak2 |
chrX:11883259-11883281 - |
TTAATGGC---TGGGA-----------------------AATAC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GT------AAA |
| droEre2 |
scaffold_4690:9691773-9691867 + |
TTAATCGT---TGGCA-----------------------AATACGTATGG------------GGTGCCACA------CAAATCGATTAGCTCTGA---ATTCGGGCGGAATTCTGAAGCACGCAT-----------------------------CCACTTTTGTA---------------GCA---------------------------TCC--------------------------------------------------------- |
| droAna3 |
scaffold_13417:2459297-2459411 - |
TAAGTTAC---TTATA-----------------------ATTAAATTTAAA-----------TATTTCGAAATAAAA--------------CTATAAAATAATTGTCTTATTTCAG------------------AAAATATGTACATATTTATACCAATCTT--------------------------------------------------------------------------------TACATATTTCAATAAAAAT------ATA |
| dp4 |
chrXL_group1e:12509670-12509797 - |
TTAAGCAT---TTTAATTTTTGTATTTTTCAAGATCGAAAATATTTATGC-GCGTAAATTTAAGAGTAAGTCCTTA-GTAAGTAAGGAAG-------------------AAC------------------------------------TACGCTCTGGATTTGAT---------------ATA---------------------------CTCGTGAAGTTAAAA----------------------------------CA------ATA |
| droPer1 |
super_14:1989251-1989367 - |
CTAAGTCA---TCGTG-----------------------CGCATCTGTTGG-----------GGAGTAAAAACTAAA--------------A-------TTAGAGTGGAAACTC--------TATTTTGAGTTGAATTTTTCCATGAAAATCTGCTACATTTTGG---------------G-T-----------------------------------------------------------------TTTAAATTTGTAT------TTT |
| droWil1 |
scaffold_181096:7961895-7962008 + |
TAGATTATGATTAGAATAAGCACACACAACACACGCACA-----------------------------------------------------------------------CACACACACACACATCTATATAT-AAATCAACCAAGAAAACA---CAATATCGAA---------------GAAA--------------------------TTAGAAAGTATATAT----------------------------------AT------ATA |
| droVir3 |
scaffold_13042:902091-902097 - |
TCAATCA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6328:2896306-2896380 - |
ATGAGAAA---GGG----------------------------------------------------CTATA------GAAATTGATGACG---------------------------------------------ATATAATCCAAAACTAAGATTACAATAATA---------------ATA---------------------------ATaataaac-----------------------------------------a------aaa |
| droGri2 |
scaffold_15081:2040284-2040418 - |
TTGTAATT---TAAGA--------------------------------------------------------------------------------------------------------------------------TGAAGATGAAGACA----------ATAATAATAATAGTGATAGGAGTTGAAGAGCTTAAATGTAAACTGATAATTGTATATACAAAGTTGAAAATGTCATTCTCTGGTTATTGGATTAAAATGAGGACACTT |