
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19455254-19455375 - | CCTTCG----CT-----------------GCTGCCGTTTGCG--TTCACACAACTGCAATTTACGGCACTGGACACGATTTTCGA-TTG----C-AACGTAAGCCAGTGGCGTAGATTGCAGATTG------TGCCGCCAACCAAGAT---ATTGTACTT |
| droSim1 | chrX_random:5079001-5079113 - | CCTTCG----CT-----------------GCTGCCGTTTGCG--TCCACACAACTGCAATATACGGCACTGGACACGATTCTCGA-CCA----C-AACGTAAGCCAGTGGCGTAGATTGCAGATTG------TGCCGCCAATC------------TACTT |
| droSec1 | super_8:1737859-1737971 - | CCTTCG----CT-----------------GCTGCCGTTTGCG--TCCACACAACTGCAATTTACGGGACTGGACACGATTCTCGA-CTA----C-AACGTAAGCCAGTGGCGTAGATTGCAGATTG------TGCCGCCAATC------------TACTT |
| droYak2 | chrX:11883747-11883862 + | TCTCCG----C--------------------TGCCCTTTGCG--TCCACACAACTGCAATCAACGGCACTGGACACGATTTCCGA-GTA----C-AACGTAAGCCAGCGGCGTAGAGTGCTGGCTG------TGCCGCCCACCT------GGATGCACTT |
| droEre2 | scaffold_4690:9691187-9691301 - | CCTCCG----C--------------------TGCCCTTTGCG--TCCACACAATTGCAATCAACGGCACTGGACACGATTTTCGA-GTA----C-AACGTAAGCCAGCGGCGTAGAATGCTGGTTG------TGCCGCCCACT-------GGATGCACTT |
| droAna3 | scaffold_13417:2460507-2460635 + | CAACTATCGGTC-----------------GCTGTAATTCGCG--TCTACACAACTGCAATCAACGTCACTGGACACGGATATCAA-CCA----C-CACGTAAGCCAGCAGCGTCGAATGCTGTCTG------TGCCGCCGAGCTTGCCCTGTCCGCACTC |
| dp4 | Unknown | ---------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer1 | super_14:1991206-1991216 + | TAGTTG----TT-----------------GTT-------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 | scaffold_181096:7961115-7961220 - | AAATCG----CC-----------------GCTAATCATTG------TACACAACTGCAATCAACGTCACTGGACACGGATATCGT-TTGTTTTT-AACGTAAGCCAGCGTCGTCGAATGCTGCCTG------TGTTGCCAA------------------- |
| droVir3 | scaffold_13042:902659-902773 + | TTCTTG----TTTGAATCAAT------ACGTTGAGATATGCA--TCAGGACAGCTCTACTCAACGGCAATGGAACCGAGTGC----GTCAAAGAGCTCGT-TCCCAAAAGCGTAGAGTACGGC-TGTGCCTG---------------------------T |
| droMoj3 | scaffold_6328:2897048-2897148 + | GTTGTG----AT-----------------G--------TGCAAACGGGGGCAGCTCTACTCAACGGCTATGGAACCGAGTGGCTTTGTATGATTGCTCGT-TCCCAAAAGCGTAGAGTACGGC-TGTACCCG---------------------------- |
| droGri2 | scaffold_15081:2040911-2041055 + | TTATTG----ATTGATTCGATTCAGTTACGTTGCGATTTGCA--TTGGGACAGCTTTACTCAGCGGCATTGAAACTAGGAGA----ATCATGTTCCTTAC-TTCCAAAAGCGCTGAATACGGC-TGGACCTGTGTTGGTTATCGAACT---GCCGTACTT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19455254-19455375 - | CCTTCG------------------CTGCTGCCGTTTGCG--TTCACACAACTGCAATTTACGGCACTGGACACGATTTTCGA-TTG----C-AACGTAAGCCAGTGGCGTAGATTGCAGATTG------TGCCGCCAACCAAGAT---ATTGTACTT |
| droSim1 | chrX_random:5079001-5079113 - | CCTTCG------------------CTGCTGCCGTTTGCG--TCCACACAACTGCAATATACGGCACTGGACACGATTCTCGA-CCA----C-AACGTAAGCCAGTGGCGTAGATTGCAGATTG------TGCCGCCAATC------------TACTT |
| droSec1 | super_8:1737859-1737971 - | CCTTCG------------------CTGCTGCCGTTTGCG--TCCACACAACTGCAATTTACGGGACTGGACACGATTCTCGA-CTA----C-AACGTAAGCCAGTGGCGTAGATTGCAGATTG------TGCCGCCAATC------------TACTT |
| droYak2 | chrX:11883747-11883862 + | TCTCC---------------------GCTGCCCTTTGCG--TCCACACAACTGCAATCAACGGCACTGGACACGATTTCCGA-GTA----C-AACGTAAGCCAGCGGCGTAGAGTGCTGGCTG------TGCCGCCCACCT------GGATGCACTT |
| droEre2 | scaffold_4690:9691187-9691301 - | CCTCC---------------------GCTGCCCTTTGCG--TCCACACAATTGCAATCAACGGCACTGGACACGATTTTCGA-GTA----C-AACGTAAGCCAGCGGCGTAGAATGCTGGTTG------TGCCGCCCACT-------GGATGCACTT |
| droAna3 | scaffold_13417:2460507-2460635 + | CAACTAT-CGG-------------TCGCTGTAATTCGCG--TCTACACAACTGCAATCAACGTCACTGGACACGGATATCAA-CCA----C-CACGTAAGCCAGCAGCGTCGAATGCTGTCTG------TGCCGCCGAGCTTGCCCTGTCCGCACTC |
| droWil1 | scaffold_181096:7961115-7961220 - | AAATCG------------------CCGCTAATCATTG------TACACAACTGCAATCAACGTCACTGGACACGGATATCGT-TTGTTTTT-AACGTAAGCCAGCGTCGTCGAATGCTGCCTG------TGTTGCCAA------------------- |
| droVir3 | scaffold_13042:902659-902773 + | TTCTTGTTTGAATCAAT------A-CGTTGAGATATGCA--TCAGGACAGCTCTACTCAACGGCAATGGAACCGAGTGC----GTCAAAGAGCTCGT-TCCCAAAAGCGTAGAGTACGGC-TGTGCCTG---------------------------T |
| droMoj3 | scaffold_6328:2897048-2897148 + | GTTGTG------------------AT--------GTGCAAACGGGGGCAGCTCTACTCAACGGCTATGGAACCGAGTGGCTTTGTATGATTGCTCGT-TCCCAAAAGCGTAGAGTACGGC-TGTACCCG---------------------------- |
| droGri2 | scaffold_15081:2040911-2041055 + | TTATTGATTGATTCGATTCAGTTA-CGTTGCGATTTGCA--TTGGGACAGCTTTACTCAGCGGCATTGAAACTAGGAGA----ATCATGTTCCTTAC-TTCCAAAAGCGCTGAATACGGC-TGGACCTGTGTTGGTTATCGAACT---GCCGTACTT |
| Species | Read pileup | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||
| dp4 |
|
| -37.8 | -37.0 |
|---|---|
 |
 |
| -34.0 | -33.5 | -33.2 |
|---|---|---|
 |
 |
 |
| -31.9 | -31.1 |
|---|---|
 |
 |
| -25.8 | -25.6 | -25.5 | -25.4 | -25.3 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
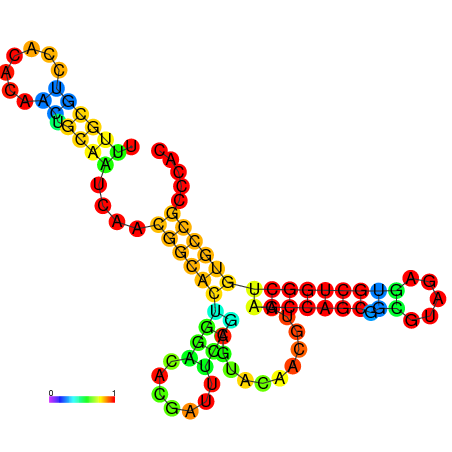 |
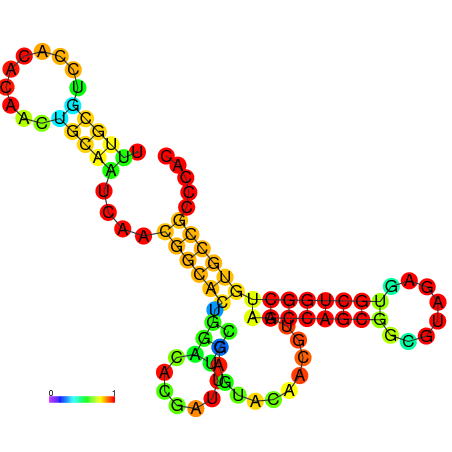 |
| -24.0 | -23.8 | -23.4 | -23.1 | -23.1 | -23.0 |
|---|---|---|---|---|---|
 |
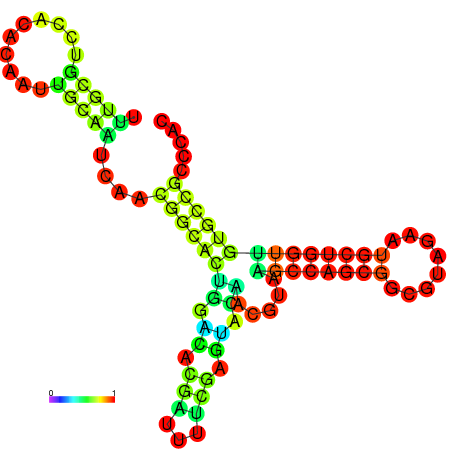 |
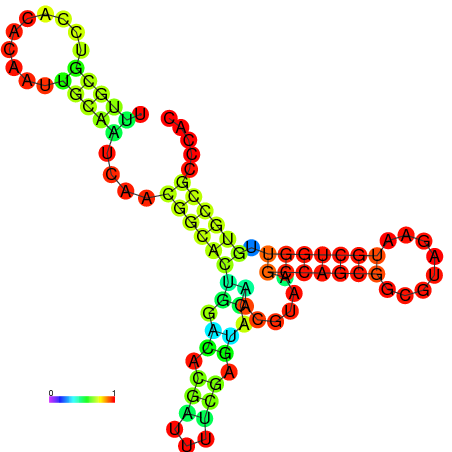 |
 |
 |
 |
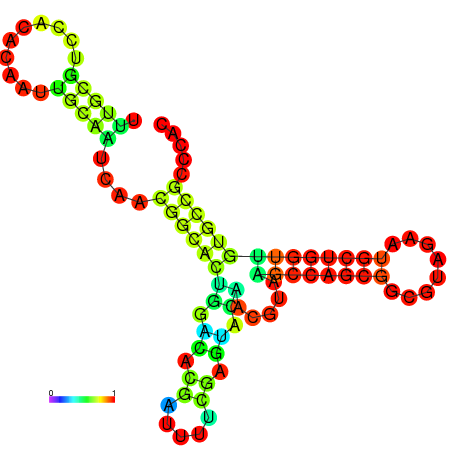
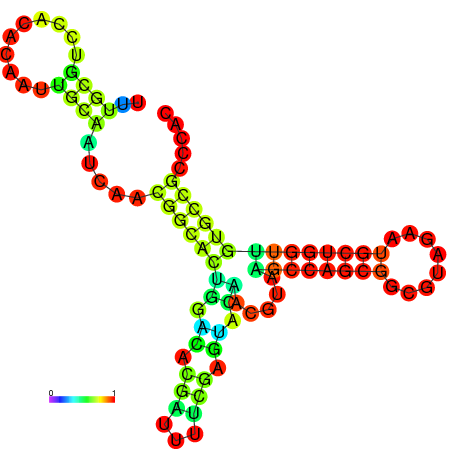
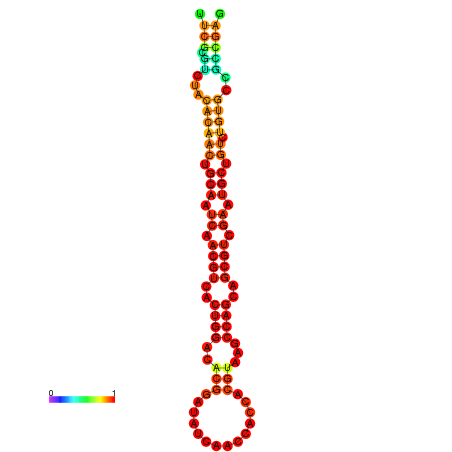
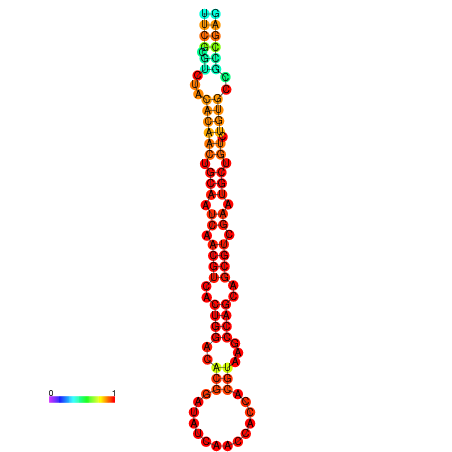
| -23.4 | -23.3 | -22.9 | -22.9 | -22.8 | -22.8 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
No secondary structure predictions returned
No secondary structure predictions returned
| -18.7 | -18.7 | -18.3 | -18.0 | -18.0 | -17.9 |
|---|---|---|---|---|---|
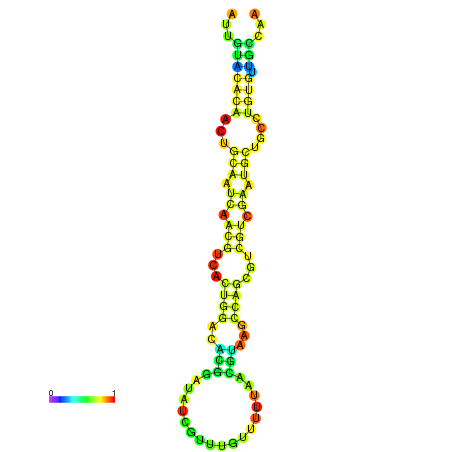 |
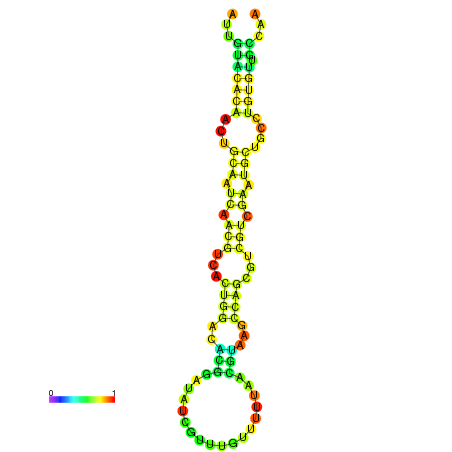 |
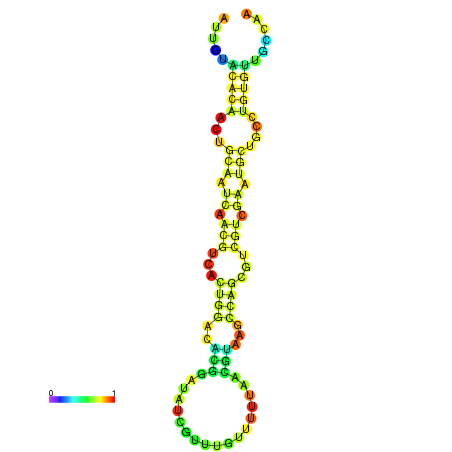 |
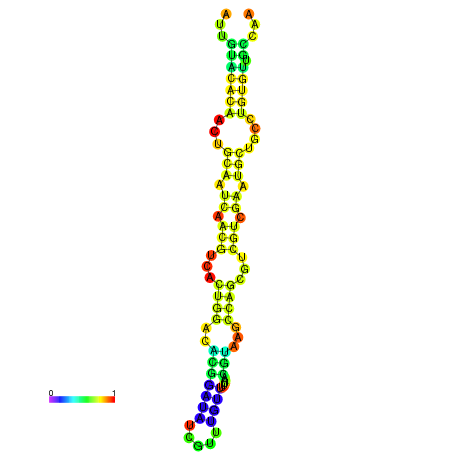 |
 |
 |
| -34.1 | -34.1 |
|---|---|
 |
 |
| -36.2 | -36.2 | -36.2 | -36.2 | -36.2 | -36.2 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -31.9 | -31.5 | -31.2 | -31.2 | -31.2 | -31.1 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
Generated: 01/05/2013 at 05:34 PM