
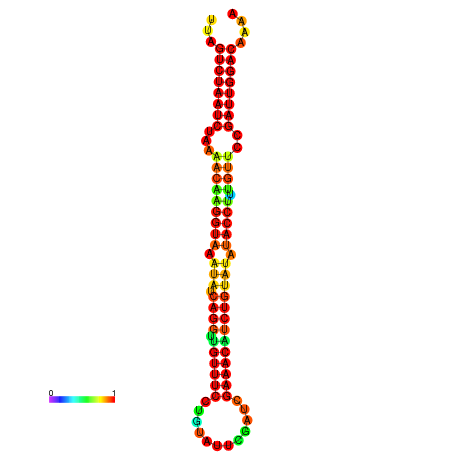
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5641975-5642093 + | TACAACTAA----------------------TTTTTCTTAGTCTAATCTAAAACAAGGTAAATATCAGGTTGTTTCCTGTATTCGA---TCGAAACATCTGTATATACCTTTGTTCCGATT-GGACA-AAAGTG--CGTCTTC--------------TA--GTT |
| droSim1 | chr2L:5435040-5435157 + | TACTACTGA----------------------TT-TTCTTAGTCTAATCTAAAACAAGGTAAATATCAGGTTGTTTCCTGTATTAGA---TCGAAACATCTGTATATACCTTTGATCCGGTT-GGACA-AAAGTG--CATCTTC--------------TA--GTG |
| droSec1 | super_5:3719693-3719811 + | TACTACTGA----------------------TTTTTCTTAGTCTAATCTAAAACAAGGTAAATATCAGGTTGTTTCCTGTATTAGA---TAGAAACATCTGTATATACCTTTGATCCGGTT-GGACA-AAAGTG--CATCTTC--------------TA--GTG |
| droYak2 | chr2L:13551185-13551307 - | CACTACTGC---A------------------TTGTTCTTGGTCTAATCTAAAACAAGGTACATATCAGGTTGTTTCCTATATTGGATGATCGAAACATCTGTATATACCTTTGTTCCGAAT-GGACG-AAAGAG--CATCTTC--------------AA--GTT |
| droEre2 | scaffold_4929:5730368-5730489 + | CAGTGCTGC----------------------TTGTTCTTAGTCTAATCTAAAACAAGGTACATATCAGGTTGTTTCCTGTATTGGATGATCGAAACATCTGTATATACCTTTGTTCCGAAT-GGACG-AAAGAG--CATCTTC--------------TA--GTT |
| droAna3 | scaffold_12916:11085426-11085560 + | TAATTA--GTTAAAAA---------------GTGTTCTTGATTTAATCTAAAACAAGGTACATATCTGGTTGTTTCCTATATTCAAAGTTTGAAACATCTGTATCTACCTTTGTTTCGATT-AAACT-AAAGAG--CTACT---AAGA---ATACGAGT--GTT |
| dp4 | chr4_group4:3181623-3181668 - | AAGAAGAGGA--AAA--------GATAATGT-GA-------------TTAATACAA-------------------------GTGGCTGATTGGA---------------------------------------------------------------------- |
| droPer1 | super_10:2200549-2200594 - | AAGAAGAGGA--AAA--------GATAATGT-GA-------------TTAATACAA-------------------------GTGGCTGATTGGA---------------------------------------------------------------------- |
| droWil1 | scaffold_180708:8625051-8625198 - | CATAGTTAAT--TGA--------GTTAATTG-AATTCTAATTCTAGTCTAAAACAAGGTACATATCAGGTTGTTTCGTATATTTGAAGCCTGAAACATCTGTATCTACCTTTGTTTCGAATAGGACT-AAAGAG--TTTTCTTTAAGCCCAACACATTT--GTT |
| droVir3 | scaffold_12963:3616326-3616465 + | TGGAGTTAATGTACGGATGGGCA--------TAAACAACAATCTAATCTGAAACAAGGTACATATCAGAGTGTTTCTTAAATTTGATTATTGAAACCTCTGTATATACCTTTCATTGGATT-AAACT-GATAAGCACATGTTT--------------CAAAAGT |
| droMoj3 | scaffold_6500:14019361-14019489 + | TGGAGTGCGC--A------------------CTG-AATCGGTCTAATCTAAAACAAGGTACATATCAGAGTGTTTCCCGTATTCGAGTTTTGAAACCTCTGTATATACCTTTCATTAGATT-CAACCGAAAGTC--CATA----AATATT---GA--TA--TAT |
| droGri2 | scaffold_15252:3148184-3148306 + | GCAAAATAA-----------------------------CAGTCTAATCCAAAACAAGGTACATATTAGAGCGTTTCCTATCTTTGAGTATTGAAACCTCTGTATATACCTTCA-TTCGATT-CGACT-GATAATCTCATT----AAAATC---ATTTCA--GTT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5641975-5642093 + | TACAACTAA--------------TTTTTCTTAGTCTAATCTAAAACAAGGTAAATATCAGGTTGTTTCCTGTATTCGA---TCGAAACATCTGTATATACCTTTGTTCCGATT-GGACA-AAAGTG--CGTCTTC--------------TA--GTT |
| droSim1 | chr2L:5435040-5435157 + | TACTACTGA--------------TT-TTCTTAGTCTAATCTAAAACAAGGTAAATATCAGGTTGTTTCCTGTATTAGA---TCGAAACATCTGTATATACCTTTGATCCGGTT-GGACA-AAAGTG--CATCTTC--------------TA--GTG |
| droSec1 | super_5:3719693-3719811 + | TACTACTGA--------------TTTTTCTTAGTCTAATCTAAAACAAGGTAAATATCAGGTTGTTTCCTGTATTAGA---TAGAAACATCTGTATATACCTTTGATCCGGTT-GGACA-AAAGTG--CATCTTC--------------TA--GTG |
| droYak2 | chr2L:13551185-13551307 - | CACTACTGC-------------ATTGTTCTTGGTCTAATCTAAAACAAGGTACATATCAGGTTGTTTCCTATATTGGATGATCGAAACATCTGTATATACCTTTGTTCCGAAT-GGACG-AAAGAG--CATCTTC--------------AA--GTT |
| droEre2 | scaffold_4929:5730368-5730489 + | CAGTGCTGC--------------TTGTTCTTAGTCTAATCTAAAACAAGGTACATATCAGGTTGTTTCCTGTATTGGATGATCGAAACATCTGTATATACCTTTGTTCCGAAT-GGACG-AAAGAG--CATCTTC--------------TA--GTT |
| droAna3 | scaffold_12916:11085426-11085560 + | TAAT--TAGTTAAAAA-------GTGTTCTTGATTTAATCTAAAACAAGGTACATATCTGGTTGTTTCCTATATTCAAAGTTTGAAACATCTGTATCTACCTTTGTTTCGATT-AAACT-AAAGAG--CTACT---AAGA---ATACGAGT--GTT |
| droWil1 | scaffold_180708:8625051-8625198 - | CATAGTTAATTGAGTTAATT---GAATTCTAATTCTAGTCTAAAACAAGGTACATATCAGGTTGTTTCGTATATTTGAAGCCTGAAACATCTGTATCTACCTTTGTTTCGAATAGGACT-AAAGAG--TTTTCTTTAAGCCCAACACATTT--GTT |
| droVir3 | scaffold_12963:3616326-3616465 + | TGGAGTTAATGTACGGATGGGCATAAACAACAATCTAATCTGAAACAAGGTACATATCAGAGTGTTTCTTAAATTTGATTATTGAAACCTCTGTATATACCTTTCATTGGATT-AAACT-GATAAGCACATGTTT--------------CAAAAGT |
| droMoj3 | scaffold_6500:14019361-14019489 + | TGGAGTGC--GC----------ACTG-AATCGGTCTAATCTAAAACAAGGTACATATCAGAGTGTTTCCCGTATTCGAGTTTTGAAACCTCTGTATATACCTTTCATTAGATT-CAACCGAAAGTC--CATA----AATATT---GA--TA--TAT |
| droGri2 | scaffold_15252:3148184-3148306 + | GCAAAATAA---------------------CAGTCTAATCCAAAACAAGGTACATATTAGAGCGTTTCCTATCTTTGAGTATTGAAACCTCTGTATATACCTTCA-TTCGATT-CGACT-GATAATCTCATT----AAAATC---ATTTCA--GTT |
| Species | Read pileup | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||
| dp4 |
|
| -26.2 | -26.2 | -26.2 | -26.2 | -26.2 | -26.2 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -24.1 | -24.1 | -23.7 | -23.7 | -23.2 | -23.2 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -25.2 | -25.2 | -25.2 | -25.2 |
|---|---|---|---|
 |
 |
 |
 |
| -23.5 | -23.5 | -23.5 | -23.5 | -23.5 | -23.5 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -22.4 | -22.4 | -22.4 | -22.4 | -22.4 | -22.4 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -19.5 | -19.5 | -19.5 | -19.4 | -19.4 | -19.4 |
|---|---|---|---|---|---|
 |
 |
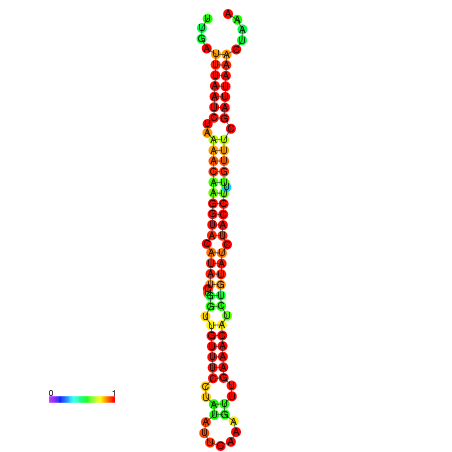 |
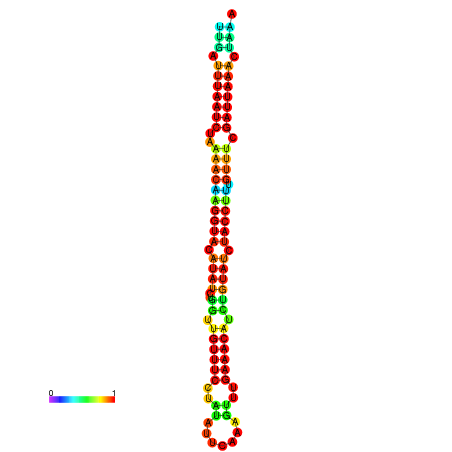 |
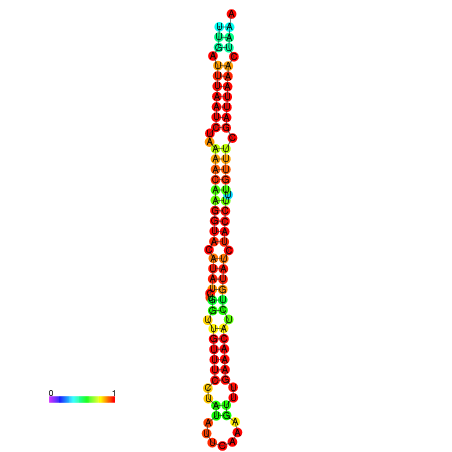 |
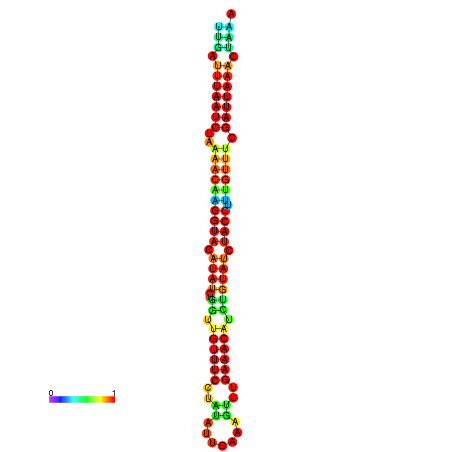 |
| -0.1 | 0.0 | 0.5 |
|---|---|---|
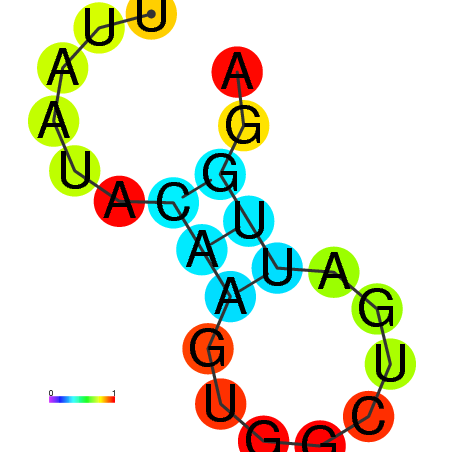 |
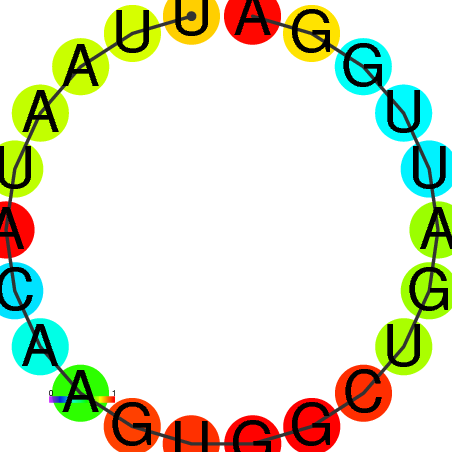 |
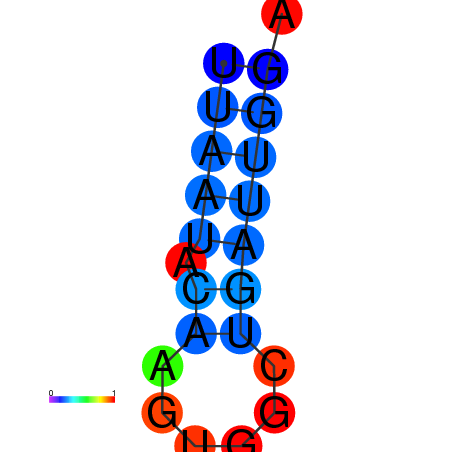 |
| -0.1 | 0.0 | 0.5 |
|---|---|---|
 |
 |
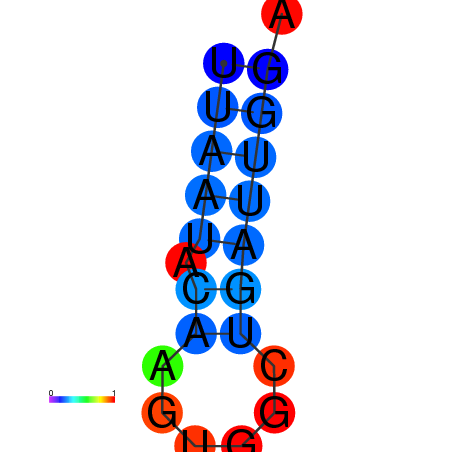 |
| -21.7 | -21.7 | -21.7 | -21.7 | -21.7 | -21.7 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -24.1 | -24.1 | -23.8 | -23.8 | -23.5 | -23.5 |
|---|---|---|---|---|---|
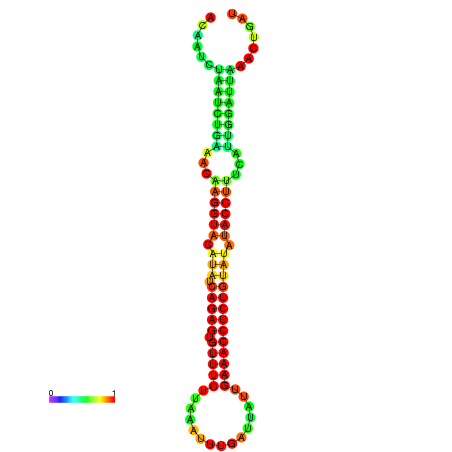 |
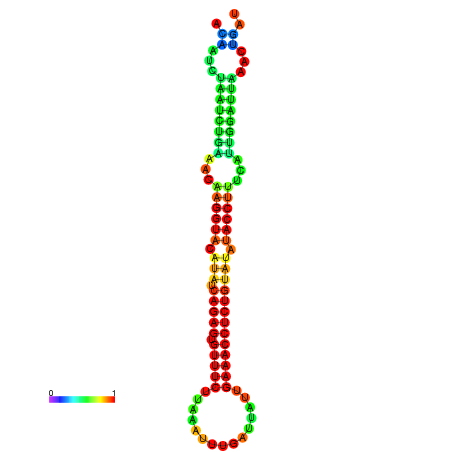 |
 |
 |
 |
 |
| -31.0 | -30.4 | -30.3 |
|---|---|---|
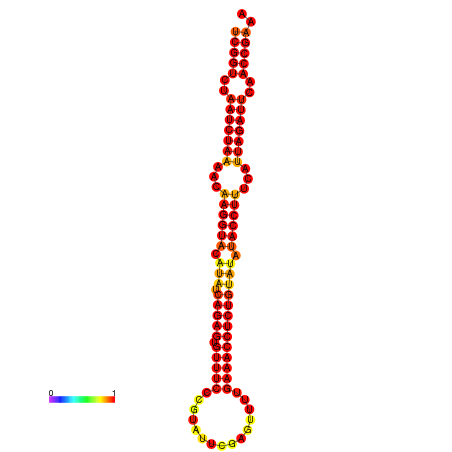 |
 |
 |
| -25.7 | -25.7 | -25.7 | -25.7 | -25.3 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
Generated: 01/05/2013 at 05:32 PM