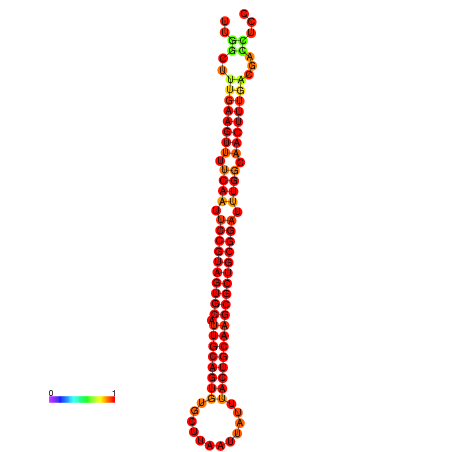
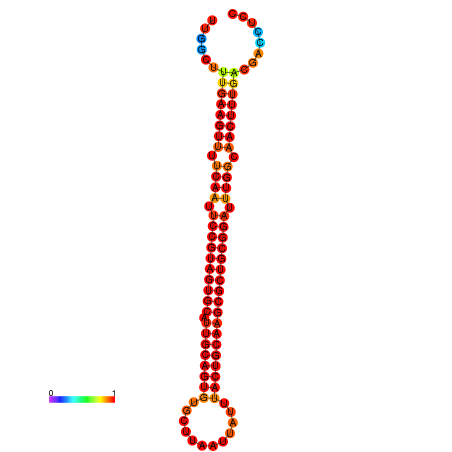
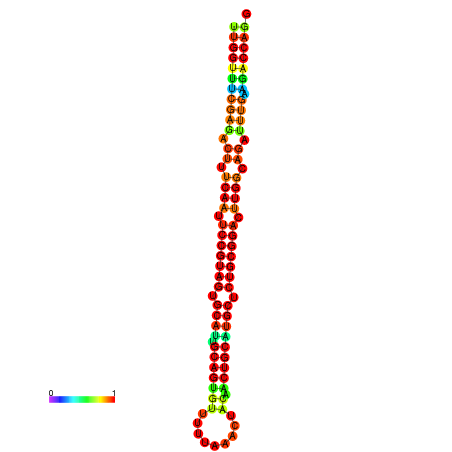
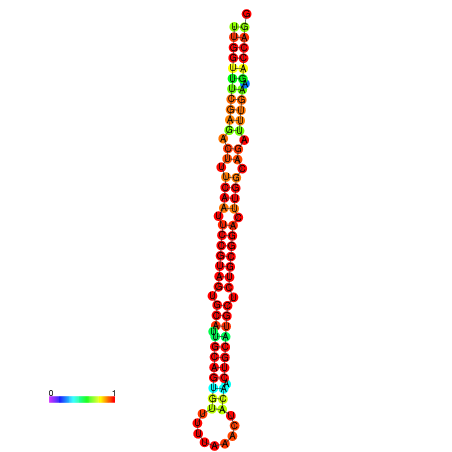
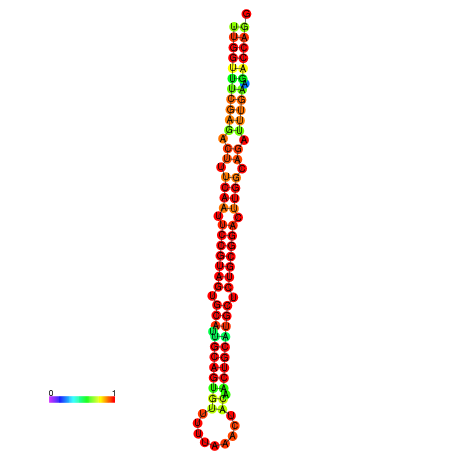
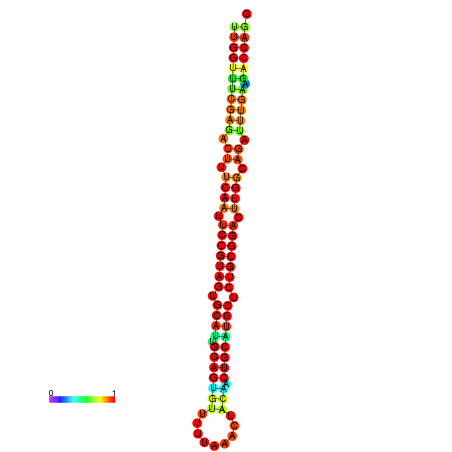
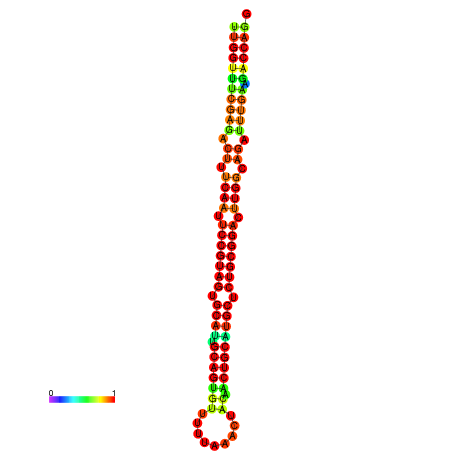
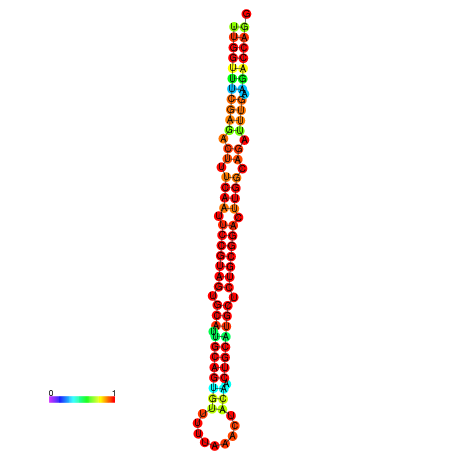
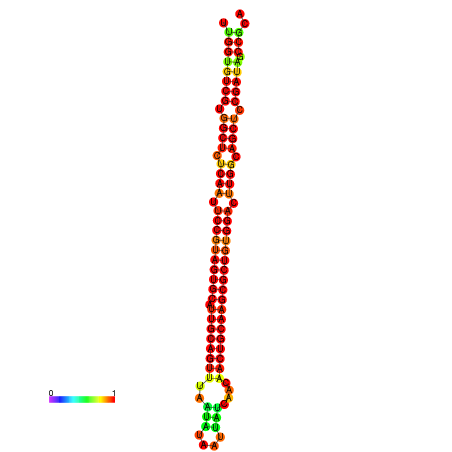
| ------------GCATTGGTGTCGTGGCTCTCAATTCCGTAGTGCATTGCAGTTTAATATAATTATCAACAACTGCAAGCGCTGTGGACTTGGCAGCTCCGATAGCCGCAAACA-CAGCTGCAGC | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ..............................TCAATTCCGTAGTGCATTGCAGT........................................................................ | 23 | 1 | 6811.00 | 6811 | 414 | 6397 |
| ..............................TCAATTCCGTAGTGCATTGCAG......................................................................... | 22 | 1 | 1955.00 | 1955 | 15 | 1940 |
| ..............................TCAATTCCGTAGTGCATTGCA.......................................................................... | 21 | 1 | 310.00 | 310 | 3 | 307 |
| .........................................................................TGCAAGCGCTGTGGACTTGGCA.............................. | 22 | 1 | 223.00 | 223 | 36 | 187 |
| ..............................TCAATTCCGTAGTGCATTGC........................................................................... | 20 | 1 | 164.00 | 164 | 1 | 163 |
| ..............................TCAATTCCGTAGTGCATTGCAGTT....................................................................... | 24 | 1 | 157.00 | 157 | 14 | 143 |
| .........................................................................TGCAAGCGCTGTGGACTTGGC............................... | 21 | 1 | 116.00 | 116 | 9 | 107 |
| ................................AATTCCGTAGTGCATTGCAGT........................................................................ | 21 | 1 | 73.00 | 73 | 8 | 65 |
| .........................................................................TGCAAGCGCTGTGGACTTGG................................ | 20 | 1 | 37.00 | 37 | 0 | 37 |
| ..............................TCAATTCCGTAGTGCATTG............................................................................ | 19 | 1 | 27.00 | 27 | 0 | 27 |
| ................................AATTCCGTAGTGCATTGCAG......................................................................... | 20 | 1 | 15.00 | 15 | 0 | 15 |
| ...............................CAATTCCGTAGTGCATTGCAGT........................................................................ | 22 | 1 | 13.00 | 13 | 1 | 12 |
| ........................................................................CTGCAAGCGCTGTGGACTTGGCA.............................. | 23 | 1 | 10.00 | 10 | 2 | 8 |
| ........................................................................CTGCAAGCGCTGTGGACTTGGC............................... | 22 | 1 | 6.00 | 6 | 2 | 4 |
| ...............................CAATTCCGTAGTGCATTGCAG......................................................................... | 21 | 1 | 4.00 | 4 | 0 | 4 |
| .............................CTCAATTCCGTAGTGCATTGCAG......................................................................... | 23 | 1 | 3.00 | 3 | 0 | 3 |
| .............................CTCAATTCCGTAGTGCATTGCAGT........................................................................ | 24 | 1 | 3.00 | 3 | 1 | 2 |
| .................................ATTCCGTAGTGCATTGCAGT........................................................................ | 20 | 1 | 3.00 | 3 | 0 | 3 |
| ................................AATTCCGTAGTGCATTGCA.......................................................................... | 19 | 1 | 3.00 | 3 | 0 | 3 |