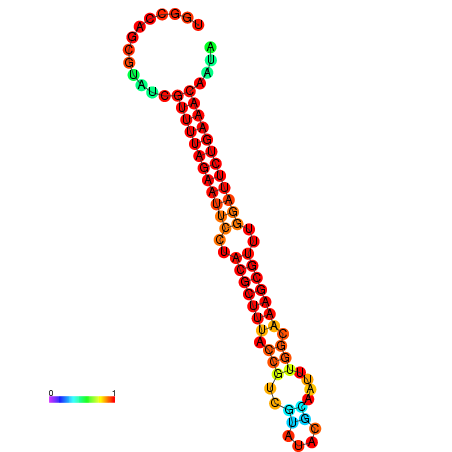
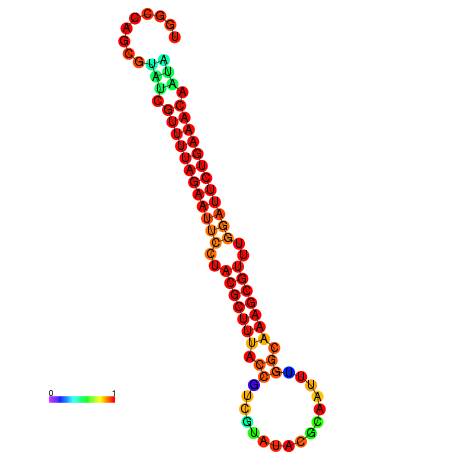
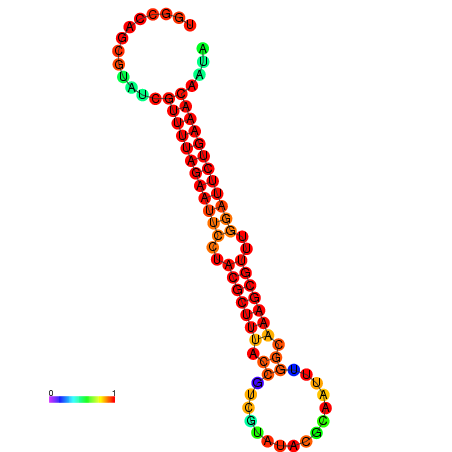
| ATAGATAGA-TAACGAGGATATTGTAGTGTTTTAGAATTCCTACGCTTTACCGTTGCAAAACGAAATTCGGCAAAGCGTTTGGATTCTGAAACCCTATGAGTCTGCATTC------AACAAAA | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ..............................TTTAGAATTCCTACGCTTTACC....................................................................... | 22 | 1 | 527.00 | 527 | 228 | 299 |
| .......................................................................CAAAGCGTTTGGATTCTGAAAC.............................. | 22 | 1 | 435.00 | 435 | 112 | 323 |
| ..............................TTTAGAATTCCTACGCTTTAC........................................................................ | 21 | 1 | 197.00 | 197 | 36 | 161 |
| ..............................TTTAGAATTCCTACGCTTTACCG...................................................................... | 23 | 1 | 26.00 | 26 | 21 | 5 |
| .......................................................................CAAAGCGTTTGGATTCTGAAA............................... | 21 | 1 | 19.00 | 19 | 11 | 8 |
| ..............................TTTAGAATTCCTACGCTTTA......................................................................... | 20 | 1 | 12.00 | 12 | 2 | 10 |
| ......................................................................GCAAAGCGTTTGGATTCTGAAAC.............................. | 23 | 1 | 12.00 | 12 | 3 | 9 |
| .......................................................................CAAAGCGTTTGGATTCTGAAACC............................. | 23 | 1 | 10.00 | 10 | 4 | 6 |
| ..............................TTTAGAATTCCTACGCTTT.......................................................................... | 19 | 1 | 8.00 | 8 | 2 | 6 |
| .......................................................................CAAAGCGTTTGGATTCTGAA................................ | 20 | 1 | 8.00 | 8 | 2 | 6 |
| ..............................TTTAGAATTCCTACGCTTTACCGT..................................................................... | 24 | 1 | 7.00 | 7 | 1 | 6 |
| .......................................................................CAAAGCGTTTGGATTCTGA................................. | 19 | 1 | 4.00 | 4 | 2 | 2 |
| ........................................................................AAAGCGTTTGGATTCTGAAAC.............................. | 21 | 1 | 2.00 | 2 | 1 | 1 |
| ....................................................GTTGCAAAACGAAATTCGG.................................................... | 19 | 1 | 2.00 | 2 | 1 | 1 |
| ......................................................................GCAAAGCGTTTGGATTCTGAA................................ | 21 | 1 | 2.00 | 2 | 1 | 1 |
| ...............................TTAGAATTCCTACGCTTTAC........................................................................ | 20 | 1 | 1.00 | 1 | 0 | 1 |
| ...............................TTAGAATTCCTACGCTTTACC....................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 |
| ................................TAGAATTCCTACGCTTTAC........................................................................ | 19 | 1 | 1.00 | 1 | 0 | 1 |
| ...........AACGAGGATATTGTAGTGT............................................................................................. | 19 | 1 | 1.00 | 1 | 0 | 1 |
| .......GA-TAACGAGGATATTGTAGTGT............................................................................................. | 22 | 1 | 1.00 | 1 | 1 | 0 |
| ..............................................................GAAATTCGGCAAAGCGTTTGGATTCT................................... | 26 | 1 | 1.00 | 1 | 0 | 1 |
| ..........TAACGAGGATATTGTAGTGT............................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 1 |