


| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:9950403-9950531 - | CCAAAGT---------CCTTT--GGCAACACATTTCATTCGCGCCTGTATCTTGCTGAACCGCTGCC-ATTATGGCCAACG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATGTGTTTAGCATTCATC----GGAGAATTT |
| droSim1 | chr2L:9739846-9739974 - | CTTAAGT---------CCTTT--GGCGGCACATTTCATTCGCGCCTGTATCTTGCTGAACCGCTGCC-ATCATGGCCAACG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATGTGTTTAGCATTCATC----GGAGGATTT |
| droSec1 | super_3:5397127-5397255 - | ACCAAGT---------CCTTT--GGCGGCACATTTCATTCGCGCCTGTATCTTGCTGAACCGCTGCC-ATCATGGCCAACG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATGTGTTTAGCATTCATC----GGAGGATTT |
| droYak2 | chr2L:12640967-12641095 - | TCCAAGT---------CCTTT--GGCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACA-ATTATGGTCTTCG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTTAGCATTCATC----GGAGGATTT |
| droEre2 | scaffold_4929:10570531-10570659 - | CACGAGT---------CCTTT--GGCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTGCC-ATTATGGCCTACG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTTAGCATTCATC----GGACGATTT |
| droAna3 | scaffold_12916:13789760-13789886 - | TG--AAT---------CCTTTCAAGCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACC-GATATGGCACAGA-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTAAGCAATCTTC----AAAAGAT-- |
| dp4 | chr4_group3:220105-220226 + | TCCTAC----------CGTGT--GCTGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTAGCC-ATAATGGTCTTTCATCCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTAAGCAATCTTC----AC------- |
| droPer1 | super_1:1694906-1695025 + | TCCTAC----------CGTGT--GCTGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTAGCC-ATAATGGTCTTTCATCCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTAAGCAATCTTC------------- |
| droWil1 | scaffold_180708:12250833-12250971 - | CCA--ATCAAAATTAACATTT-GGCCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACCAATTGTGATCTTCATTTCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTGACGGTAACATC----GTTGGAATA |
| droVir3 | scaffold_12723:3996238-3996344 + | CTA---T---------CCCTT-GGCCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACA-ATTGAGACATCTAAATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGT------------------------- |
| droMoj3 | scaffold_6500:14729857-14729979 - | -----AT---------CCTTA--GCCGGCATCTTTCATTCGCGCCTGTATCTTGCTGAACCGTTACA-ATTGTCACATCCAAATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTAGCGGTCTCATC----AAAGCA--- |
| droGri2 | scaffold_15252:3661296-3661426 - | TT---AT---------CCTTG--ACCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACC-ATTGTGACATCCAAATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTAGCGGCTTCATCACTTGGTGGATAG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:9950403-9950531 - | CCAAAGT---------CCTTT--GGCAACACATTTCATTCGCGCCTGTATCTTGCTGAACCGCTGCC-ATTATGGCCAACG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATGTGTTTAGCATTCATC----GGAGAATTT |
| droSim1 | chr2L:9739846-9739974 - | CTTAAGT---------CCTTT--GGCGGCACATTTCATTCGCGCCTGTATCTTGCTGAACCGCTGCC-ATCATGGCCAACG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATGTGTTTAGCATTCATC----GGAGGATTT |
| droSec1 | super_3:5397127-5397255 - | ACCAAGT---------CCTTT--GGCGGCACATTTCATTCGCGCCTGTATCTTGCTGAACCGCTGCC-ATCATGGCCAACG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATGTGTTTAGCATTCATC----GGAGGATTT |
| droYak2 | chr2L:12640967-12641095 - | TCCAAGT---------CCTTT--GGCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACA-ATTATGGTCTTCG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTTAGCATTCATC----GGAGGATTT |
| droEre2 | scaffold_4929:10570531-10570659 - | CACGAGT---------CCTTT--GGCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTGCC-ATTATGGCCTACG-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTTAGCATTCATC----GGACGATTT |
| droAna3 | scaffold_12916:13789760-13789886 - | TG--AAT---------CCTTTCAAGCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACC-GATATGGCACAGA-ATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTAAGCAATCTTC----AAAAGAT-- |
| dp4 | chr4_group3:220105-220226 + | TCCTAC----------CGTGT--GCTGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTAGCC-ATAATGGTCTTTCATCCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTAAGCAATCTTC----AC------- |
| droPer1 | super_1:1694906-1695025 + | TCCTAC----------CGTGT--GCTGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTAGCC-ATAATGGTCTTTCATCCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTTAAGCAATCTTC------------- |
| droWil1 | scaffold_180708:12250833-12250971 - | CCA--ATCAAAATTAACATTT-GGCCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACCAATTGTGATCTTCATTTCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTGACGGTAACATC----GTTGGAATA |
| droVir3 | scaffold_12723:3996238-3996344 + | CTA---T---------CCCTT-GGCCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACA-ATTGAGACATCTAAATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGT------------------------- |
| droMoj3 | scaffold_6500:14729857-14729979 - | -----AT---------CCTTA--GCCGGCATCTTTCATTCGCGCCTGTATCTTGCTGAACCGTTACA-ATTGTCACATCCAAATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTAGCGGTCTCATC----AAAGCA--- |
| droGri2 | scaffold_15252:3661296-3661426 - | TT---AT---------CCTTG--ACCGGCATATTTCATTCGCGCCTGTATCTTGCTGAACCGTTACC-ATTGTGACATCCAAATCCGGTTGAGCAAAATTTCAGGTGTGTGAGAAATTTGTAGCGGCTTCATCACTTGGTGGATAG |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| -41.2 | -41.1 | -40.8 | -40.7 | -40.7 | -40.6 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -41.1 | -41.0 | -41.0 | -40.9 | -40.7 | -40.6 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -41.1 | -41.0 | -41.0 | -40.9 | -40.7 | -40.6 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
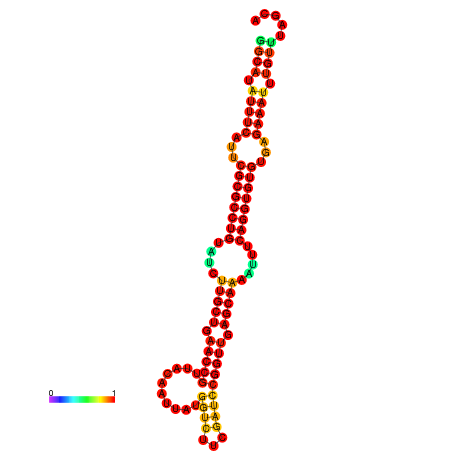
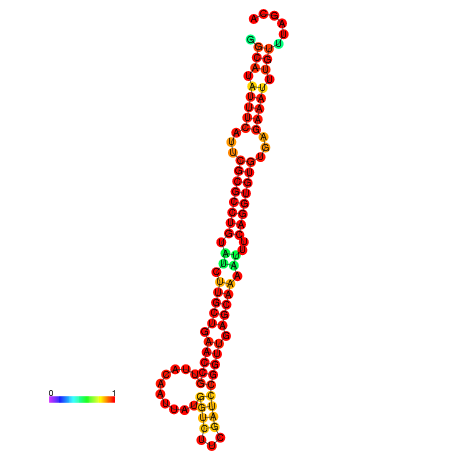
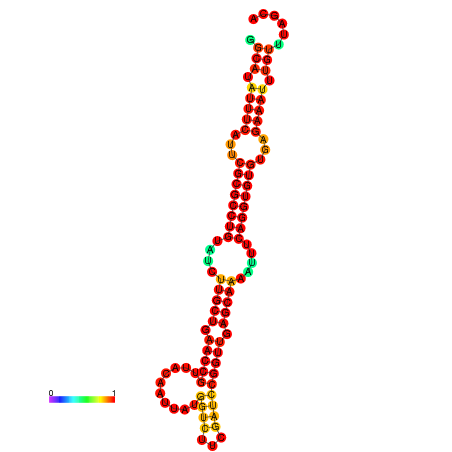
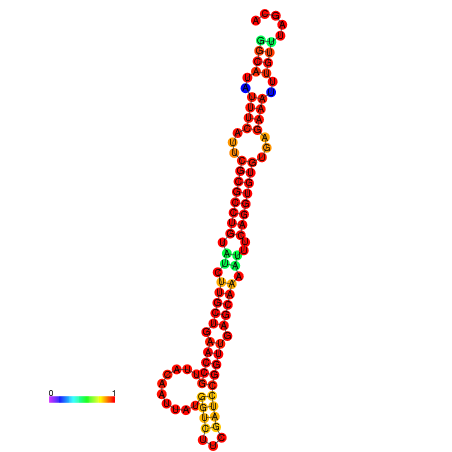
| -32.9 | -32.8 | -32.8 | -32.7 | -31.9 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| -35.3 | -35.2 | -35.2 | -35.1 | -34.9 | -34.8 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -31.8 | -31.7 | -31.7 | -31.6 | -31.6 | -31.5 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -33.8 | -33.7 | -33.5 | -33.4 | -33.4 | -33.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -33.8 | -33.7 | -33.5 | -33.4 | -33.4 | -33.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -31.8 | -31.8 | -31.7 | -31.7 | -31.2 | -31.1 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -31.5 | -31.4 | -30.8 | -30.7 | -30.7 | -30.6 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -30.0 | -29.9 | -29.4 | -29.4 | -29.3 | -29.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -32.0 | -31.9 | -31.7 | -31.6 | -31.1 | -31.0 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
Generated: 01/05/2013 at 05:30 PM