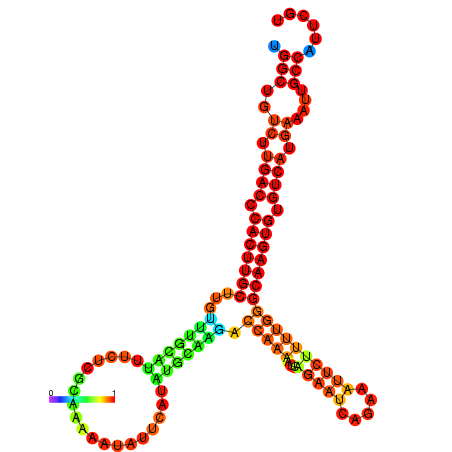
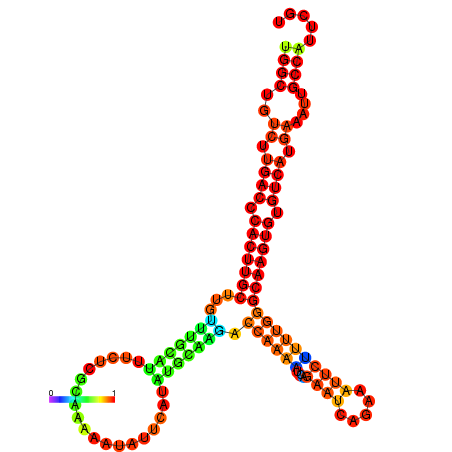
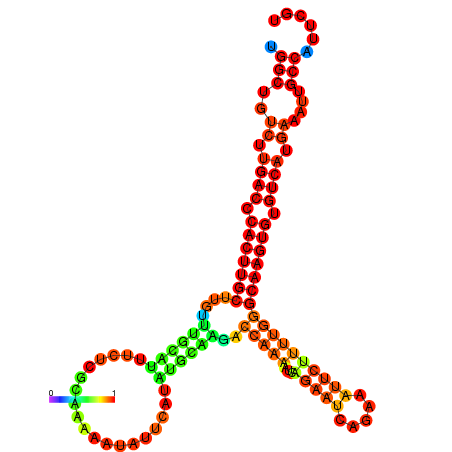
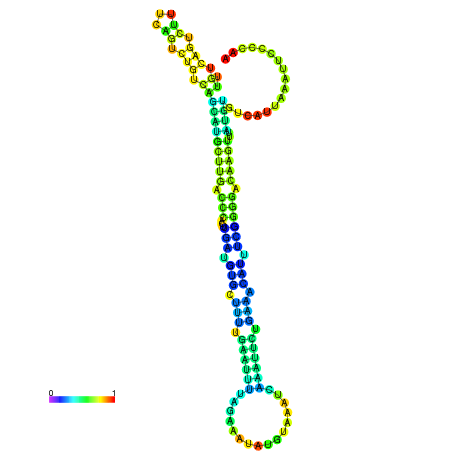
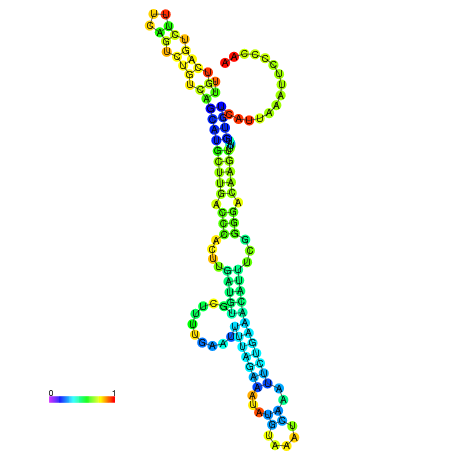
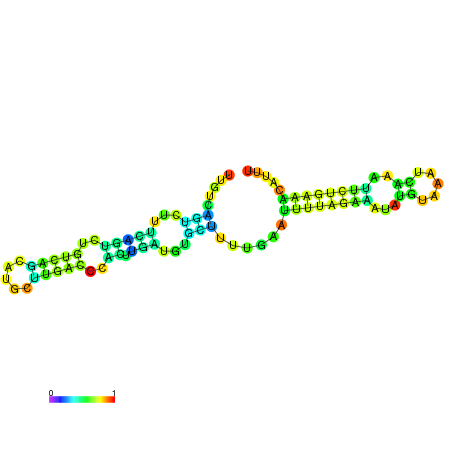
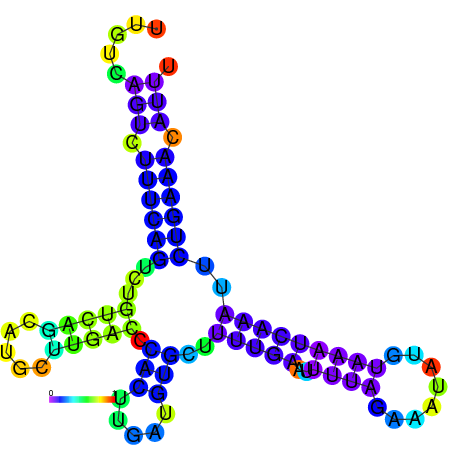
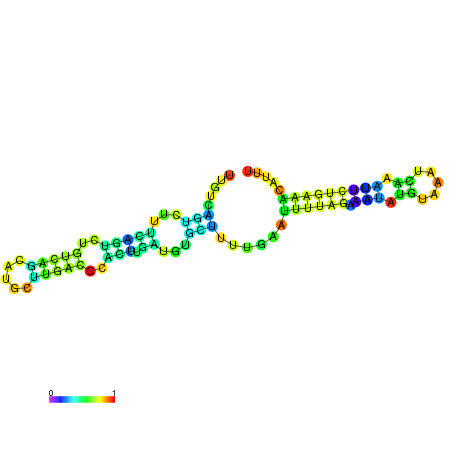
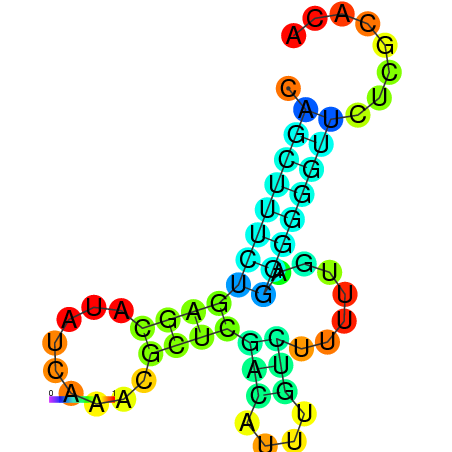
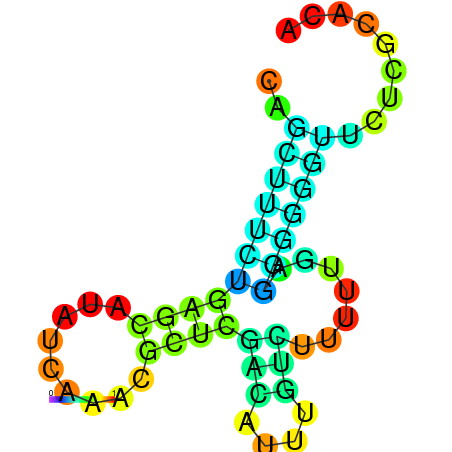
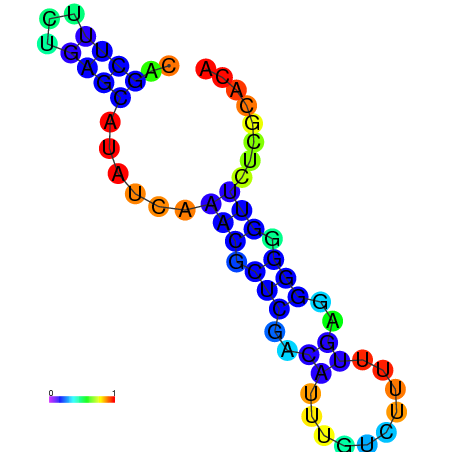
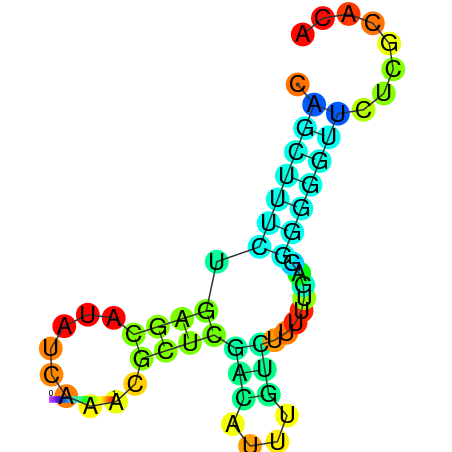
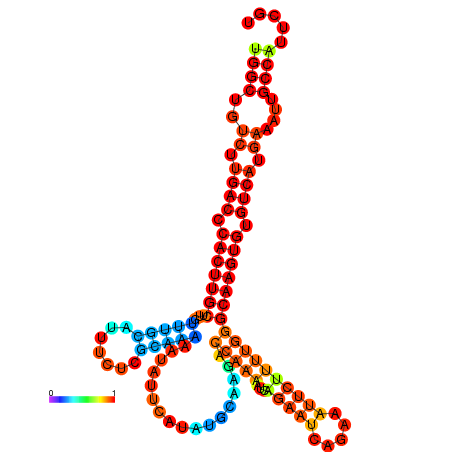| dm3 |
chr2R:15524903-15525041 + |
AA--CACATCTGCCCGCTGG-CTGGCTG------T---------CTTGACCCACTTGCT---CG---TTTGCA------TT-TCT--GAAAAATGT--ATACGCAA-----AAACCAAAATCAGAAATTCTTTTGA-GCAAG----TGTGTCATGAAATTGCC-ATTCGTTTCTGCTCTATAT-------------AC |
| droSim1 |
chr2R:14228631-14228769 + |
AA--CACATCTGCCCGCTGG-CTGGCTG------T---------CTTGACCCACTTGCT---CG---TTTGCA------TT-TCT--GAAAAATGT--ATACGCAA-----AAACCAAAATCAGAAATTCTTTTGA-GCAAG----TGTGTCATGAAATTGCC-ATTCGTTTCTGCTCTATAT-------------AC |
| droSec1 |
super_1:13039564-13039702 + |
AA--CACATCTGCCCGCTGG-CTGGCTG------T---------CTTGACCCACTTGCT---TG---TTTGCA------TT-TCT--GAAAAATGT--ATACGCAA-----AAACCAAAATCAGAAATTCTTTTGA-GCAAG----TGTGTCATGAAATTGCC-ATTCGTTTCTGCTCTATAT-------------AC |
| droYak2 |
chr2R:13649050-13649195 - |
AA--CACATCTGCCCGCTGG-CTGGCTG------T---------CTTGACCCACTTGCT---CG---TTTGCATTTGTATTTTCT--GAAAAATGT--ATACGCAA-----AAACCAAAATCAGAAATTCTTTTGA-GCAAG----TGTGTCATGAAATTGCC-ATTCGTTTCTGCTCTATAT-------------AC |
| droEre2 |
scaffold_4845:9723500-9723642 + |
AA--CACATCTGCCCGCTGG-CTGGCTGGCTG--T---------CTTGACCCACTTGCT---CG---CTTGCA------TT-GCT--GAAAAATGT--ATACGCAA-----AAACCAAAATCAGAAATTCCTTTGA-GCAAG----TGTGTCATGAAATTGCC-ATTCGTTTCTGCTCTATAT-------------AC |
| droAna3 |
scaffold_13266:9754397-9754555 - |
AAAACACAAACGCCCGCTGG-----CTG------T---------CTTGACCCACTTGCT---TG---TTTGCA------TT-TCTCGCAAAAATATTCATATGCAAGACCAAAATCAGAATCAGAAATTCTTTTGG-GCAAG----TGTGTCATGAAATTGCC-ATTCGTTTCTGCTCTCTATATGTACATCTAGTAC |
| dp4 |
chr3:12652733-12652881 + |
AA--CACGTCTCTCCGTTTGTCAGTCTTTCAGTCTGTCAGCATGCTTGACCCACTTGATGTGCT---TTTGAA------TT-TTA--GAAATATGT---------------AAATCAAATTCTGAAACATTTCGGGGACAAGTTTATGTGTCATTAAATTCCCCA----ATTTTATACTAAA---------------- |
| droPer1 |
super_2:8929657-8929759 - |
AA--CACTTCTCTCCGTTTGTCAGTCTTTCAGTCTGTCAGCATGCTTGACCCACTTGATGTGCT---TTTGAA------TT-TTA--GAAATATGT---------------AAATCAAATTCTGAAACATTT------------------------------------------------------------------ |
| droWil1 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droVir3 |
scaffold_12875:4153222-4153295 - |
AA--CACTTTTGAGTGCCAG-CTTTCTG------A---------GCATATC-AAACGCT---CGACATTTGTC------TT-TTT--GAGGGGGGT--TCTCGCAC-----A-------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6496:20896701-20896727 + |
AA--CACTTTTGACTGCTGC-CAGCCTG------C---------C--------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |