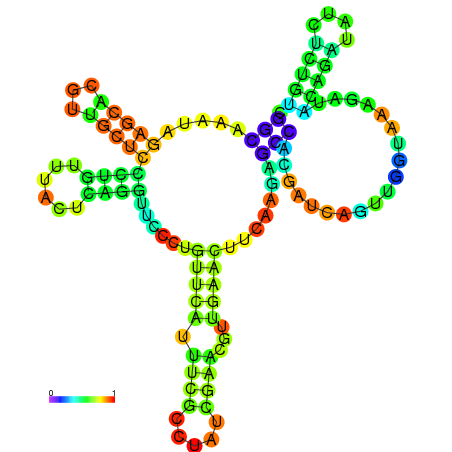
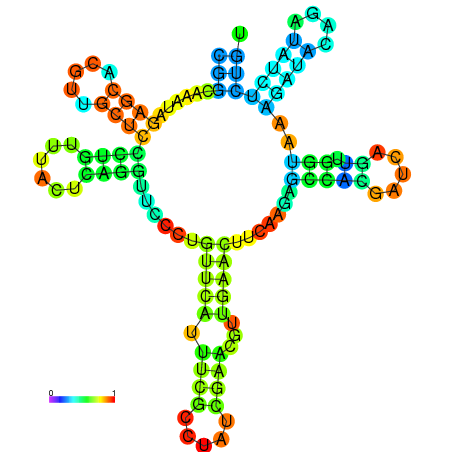
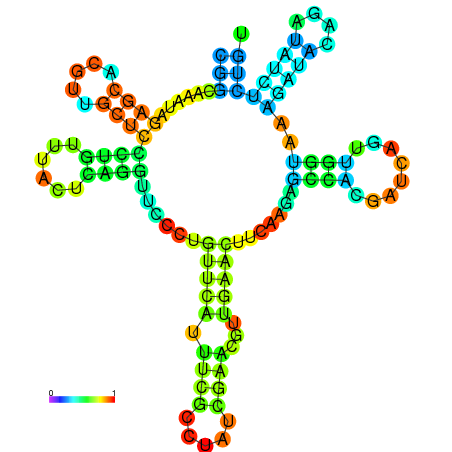
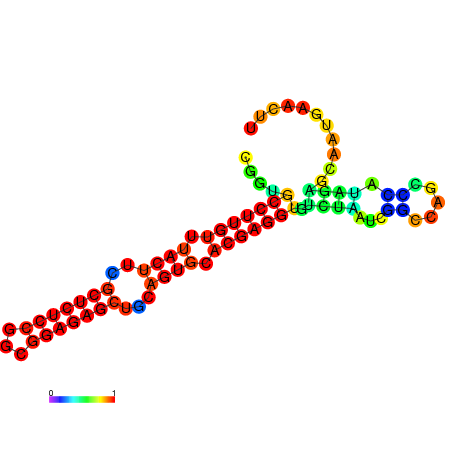
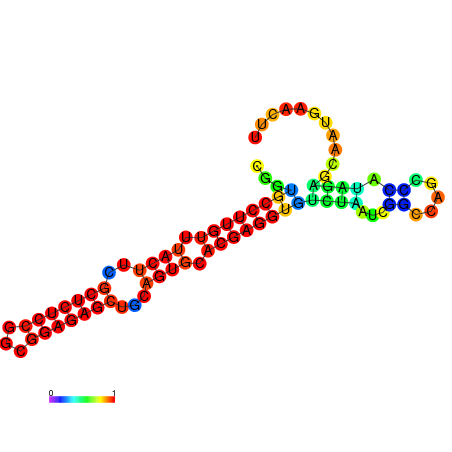
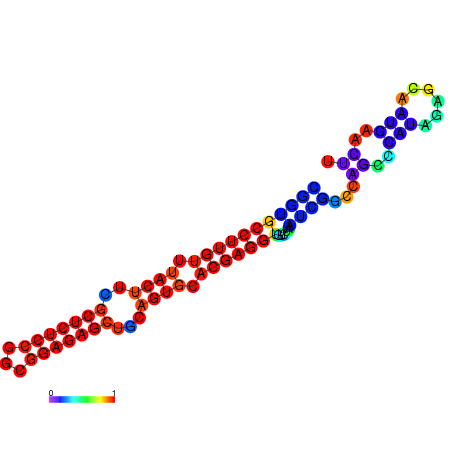
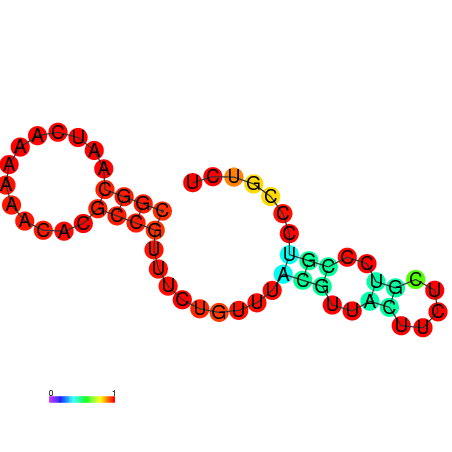
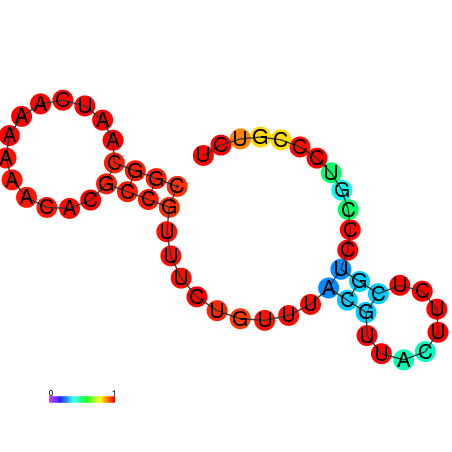
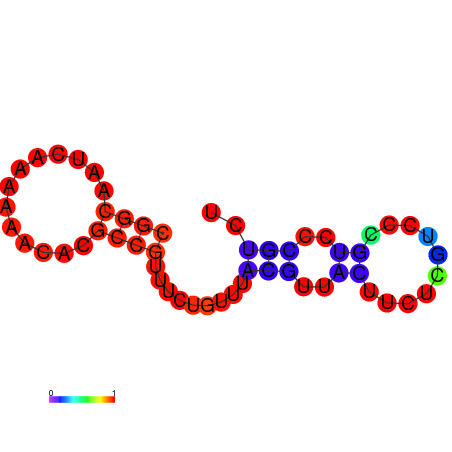
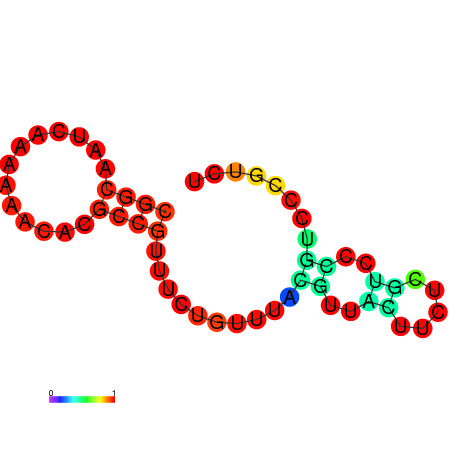
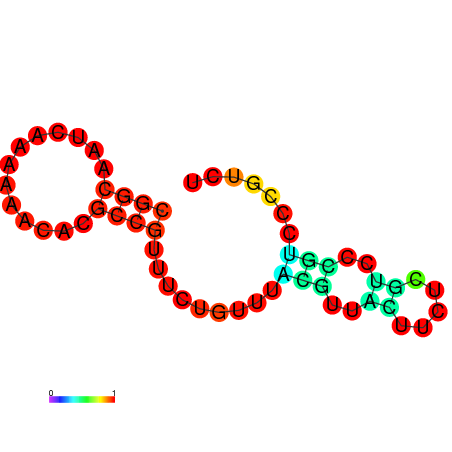
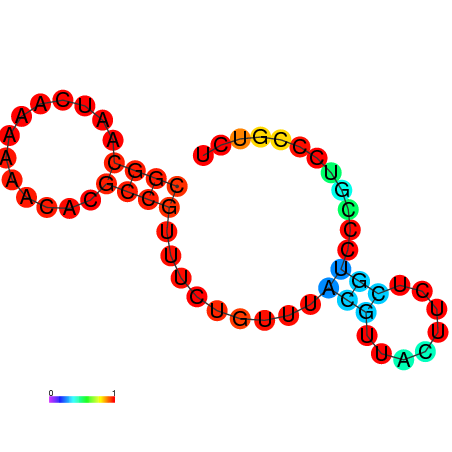
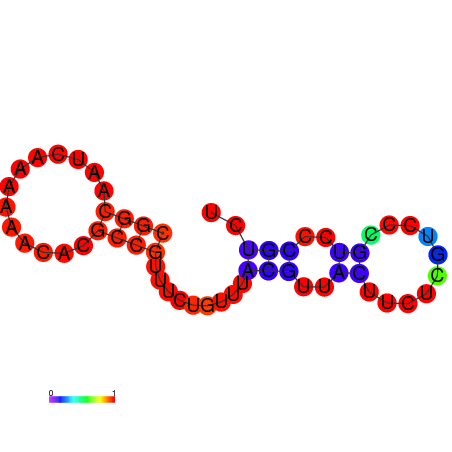
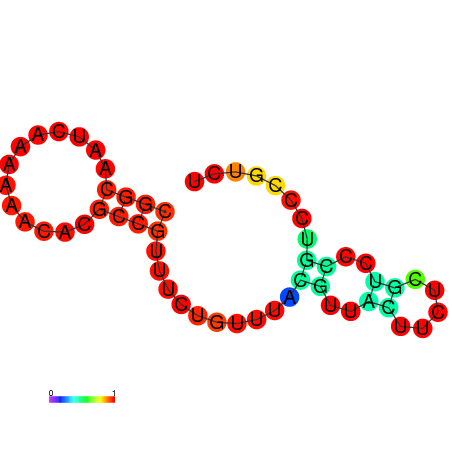
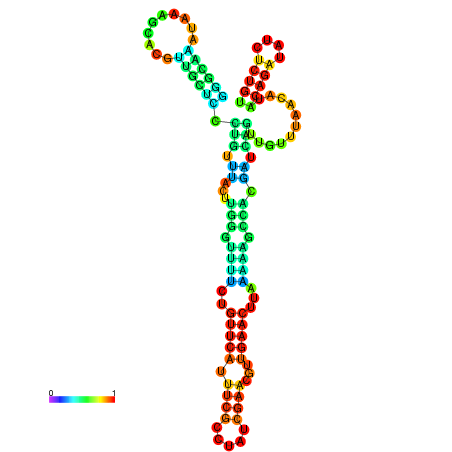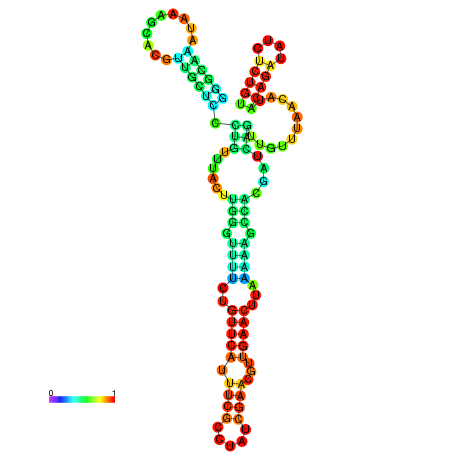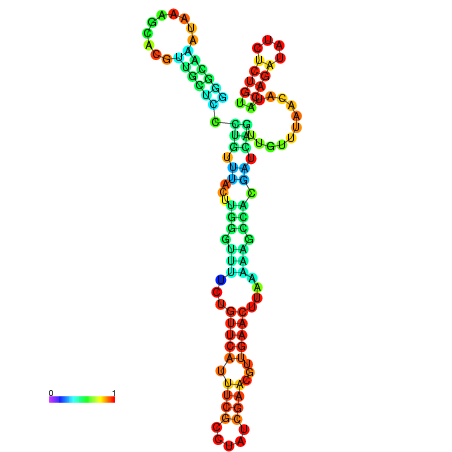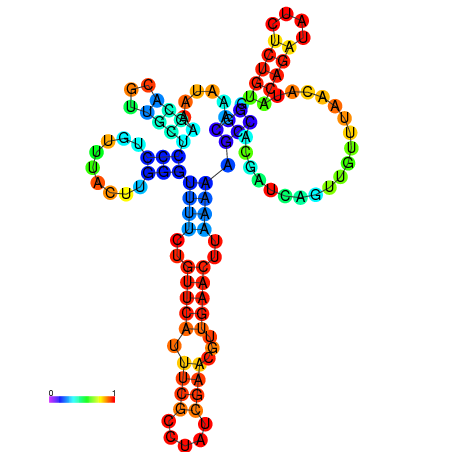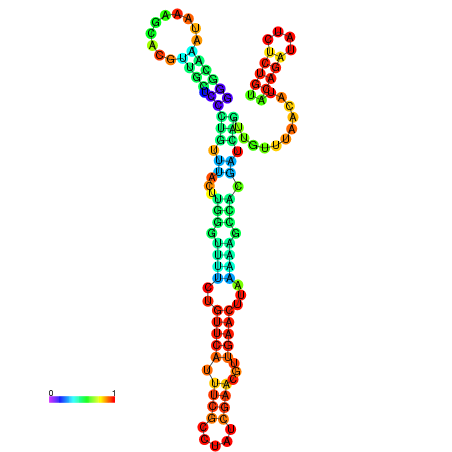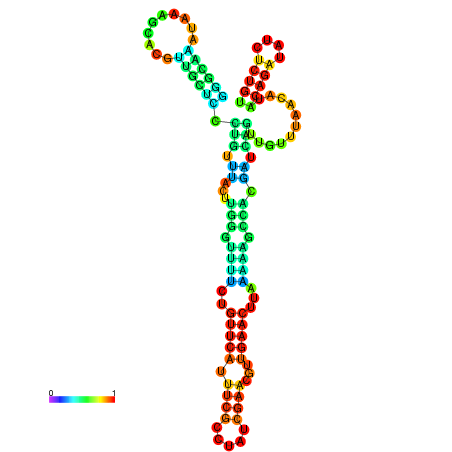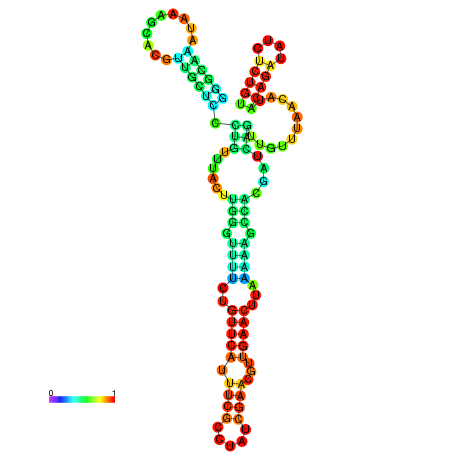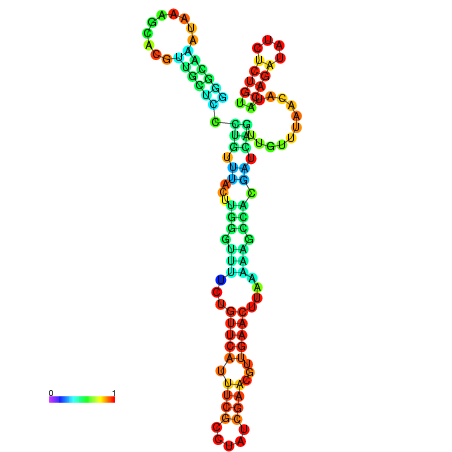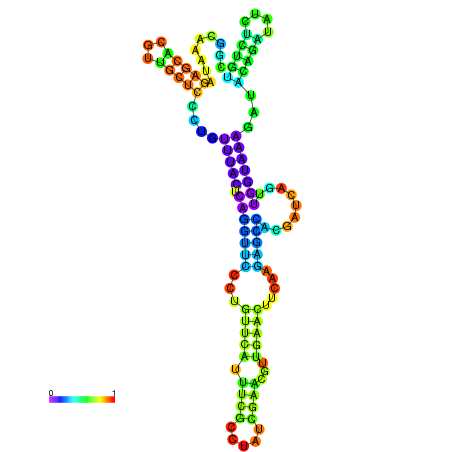| dm3 |
chr2R:2594652-2594793 - |
TAGAACTGGAAATCTGGGCAAATAA--AGCACGTTGCTCCCTGTTTACTTGGGTTTTCTG----------------------------TTCATTTCGCCTATCGAACGTTGAACTTAAAAAGCCACGATCAGTTGTTTAACATACAGATATCTCTGTACACAAGAAGATTCA |
| droSim1 |
chr2R:1414726-1414867 - |
TAGAACTGAAAGTCTCGGCAAATAG--AGCACGTTGCTCCCTGTTTACTCAGGTTCCCTG----------------------------TTCATTTCGCCTATCGAACGTTGAACTTCAAGAGCCACGATCAGTTGGTAAAGATACAGATATCTCTGTATACTAGAAGATACA |
| droSec1 |
super_1:259050-259190 - |
TAGAACTGGAAGTCTCGGCAAATAA--AGCACGTTGCTCCCTGTTTACTCAGGTTCCCTG----------------------------TTCAGTTCGCCTATCGAACGTTGAACTTCAAGAGCCACGATCAGTTGGT-AGGATACAGATATCTCTGTATACTAGAAGATACA |
| droYak2 |
chr2L:15337545-15337682 - |
TAGAAATGAAAATCTCGGCAAGTAA--AGCACGTTGCTCCATGTTTACTCAGGTTTGCTG----------------------------TTTATATCGCCTATCAAACGTTGAACTTGA---ATCACGATCAGTTGGTCAACATACATATATTTGT-CACACTAGGTGATCCA |
| droEre2 |
scaffold_4929:19950710-19950840 + |
TAGGACTGCAGATCCCGGCAACTAA--AGCACGTTGCTCCCAGTTTACTCAGGTTCGCTG----------------------------TTTATATCGCCTATCGAGCGTTGAACTTGAGGAGCCACGATCAGTTGGTCAGCATACATATA-----------AAGATGATCCA |
| droAna3 |
scaffold_13266:18414789-18414883 + |
AAGTCTA----ATTTCGGT------------------GCCTTGTTTACTTCGCTCTCCGGCGGAGAGCTGCAGTGCACGAGGTGTCTAATCGGCCAGCCCATAGAGCAATGAACTT-------------------------------------------------------- |
| dp4 |
chr3:859167-859237 - |
TAGCACAGAAAATCTCGGCAATCAAAAAACACGCCGTT-TCTGTTTACGTTACTTCTCGT----------------------------CCCGTCCCGTCT------------------------------------------------------------------------ |
| droPer1 |
super_2:1035040-1035110 - |
TAGCACAGAAAATCTCGGCAATCAAAAAACACGCCGTT-TCTGTTTACGTTACTTCTCGT----------------------------CCCGTCCCGTCT------------------------------------------------------------------------ |
| droWil1 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |