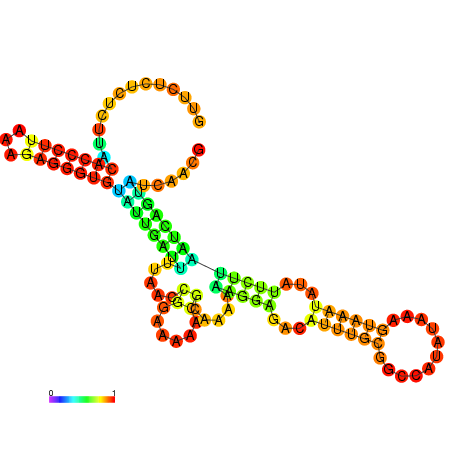
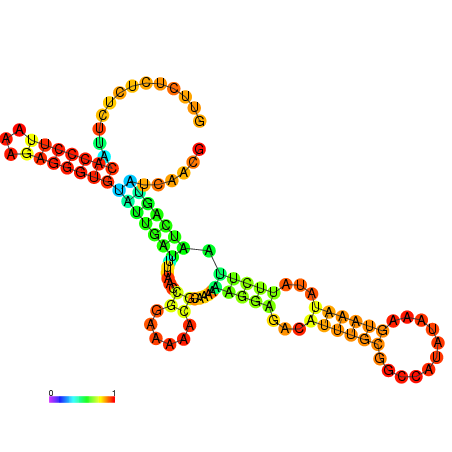
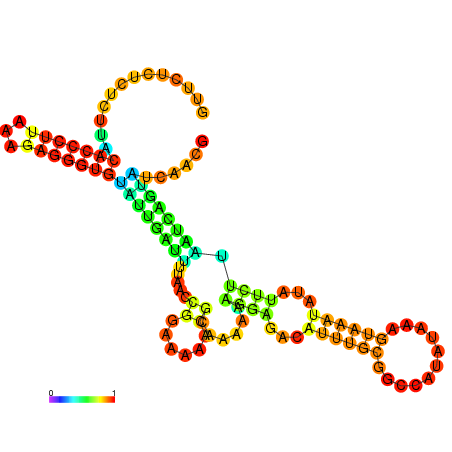
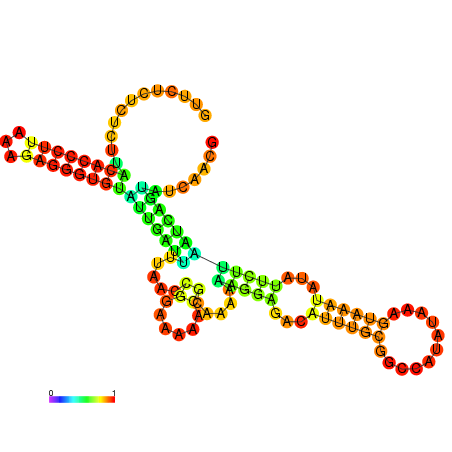
| dm3 |
chr2L:21263917-21264049 + |
GCATTGACTTCTTTTGTTCTT---TGTTTTTATTCCCTGTCAG--AGAGTATATT--GATTTTAGGTGG-----------------------AC-------ATTTGCGACCATATAAAATAT-ATATATTCCTGATAAAGAATATTTA--------ATATATCTTCTTTTTCGGTATAC |
| droSim1 |
chr2L:20861453-20861585 + |
GCACTGACTTCTTTTGTTCTT---TGTTTTTATTCCCTAGCAG--AGAGTATATT--GATTTTAGGCGG-----------------------TC-------ATTTGCGACCATATAAAGTAT-ACATATTCCTGATCAGAACTATTTA--------ATATATCTTTTTTTTCGGTACAC |
| droSec1 |
super_18:1195266-1195398 + |
GCACTGACTTCTTTTGTTCTT---TGTTTTTATTCCCTAGCAG--AGAGTATATT--GATTTTAGGCGG-----------------------TC-------ATTTGCGACCATATAAAGTAT-ACATATTCCTGATCAGAACTATTTA--------ATATATCTTTTTTTTCGGTATAC |
| droYak2 |
chrU:19793626-19793766 + |
GAACTTAATTATATCGTTCTC---T-CTCTTACACCCTTAAAG--AGGGTGTATT--GATTTTAACCGGAAAAAC-----GCAAAAAAGGAGAC-------ATTTGCGGCCATATAAAGTAA-ATATATTCTTAATCAGTATCAA-----------------CGTCAGTTTTAAGATCC |
| droEre2 |
scaffold_4784:24793627-24793751 + |
TAATTGAATTTCTTTGGTTTT---T-TTACCATTTACTTTTAG--AGAATATACTAGAATTTC--TGAGAAGTGTGTAACAGGCAGAAGGAAAC-------GTTTTCGACCATATAAATTAT-ATATATTCTTGATCAGGA-------------------------------------- |
| droAna3 |
scaffold_13499:388035-388162 - |
TTATT----TCCTTTGTTCTT---CGTTTTTATACCCTTGCAG--AGGGTATTAT--AATTTTGGTCAAAAGTGTGCAACGCAGAGAAGGAGAC-------ATCTCCGACCCTATAAAGTAT-ATATATTCTTGATCAGGATCACCT-------------------------------- |
| dp4 |
chr2:13223937-13224089 - |
CCGTTCGCCTTTTTGGTTCTTTATTGTTTTTATACCCTTCTAG--AGAGTATTAC--GATTTTGGGCTAATGTCTGCAAGACTGGAAC--------CTGCAACATAGAATCCCATAGGGTATTATATAT-----ATGAGGATCGGTAG--------CCATCTCT-CTTTTTCTATACAC |
| droPer1 |
super_0:7229185-7229317 + |
CCGTTCGCCTTTTTGGTTCTTTATTGTTTTTATACCCTTCTAG--AGAGTATTAT--GATTTTGGGCTGCTGTCTGCAAGGCTGAAAT--------CTATTGCATAGAATCCTATAGGGTATTATATAT-----GTCAGGA-----------------------------TCGGTAGCC |
| droWil1 |
scaffold_181150:776667-776791 - |
T--------TTTTTTGATTTT---CTTTTTTATAACCTTGCAAAAAGGGTATATT--CATTTTGGTCAAAGGAAT-----------------GC-------ATCTCCGACCATGTAAATTAT-ATATATTCTTGATTAGCACGACGAGACGAGTTCATATAG----------------C |
| droVir3 |
scaffold_12963:16876342-16876466 - |
GAACTCGTTTCTGGTGTACGT-CGTGTTTTTATACCCTTGTAG--AGAGTATTTT--AATTTTGTCGTGGAATGTGCAACGCATGGATGGAGAC-------ATTTGGGACCCCATAAAGTAT-ATGTATTCTAGATAA----------------------------------------- |
| droMoj3 |
Unknown |
GGTTTCAGTTCTTTTGTTTTT---TTTTG--ATATCCCAAA-------------------------------TGTGAAGTGCTAAAGAGTTGAAATTCGGAATGTAGATTGGCAT-----------------TGAGTTCAATAAGATA--------TGGCGGATATATTGTATAC---A |
| droGri2 |
scaffold_3049:4191-4318 - |
ATTGACAAAT---TTATTTTT---AATTCTTCTTTTGTTCGAG--TGGGTATAAC--AGGTAGAAGGAA-----------------------GC-------ATCTCCGACGCCATAAAGTAT-ATATGTTTCTGAGAAGGATCAATTG--------AGGAGTCGATGTAGTTCGT---C |