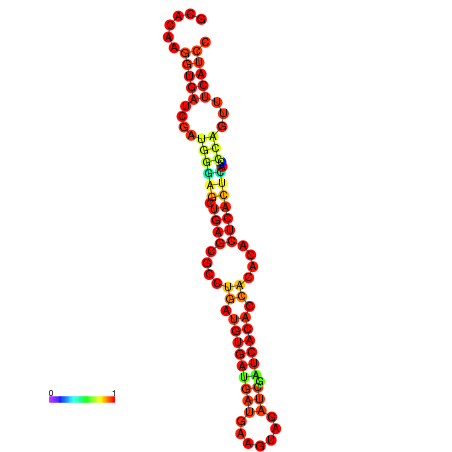
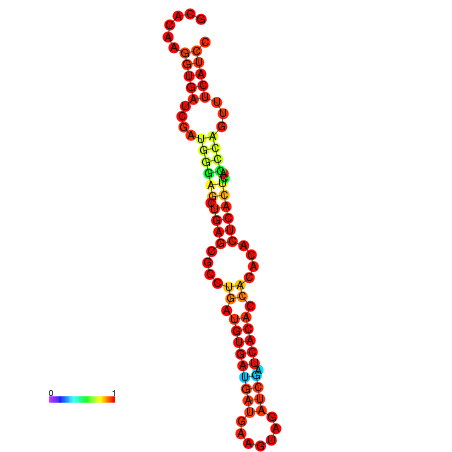
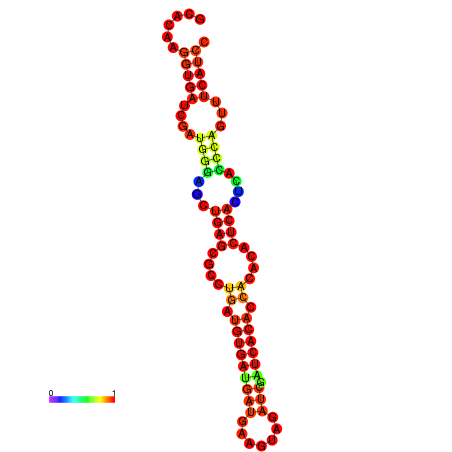
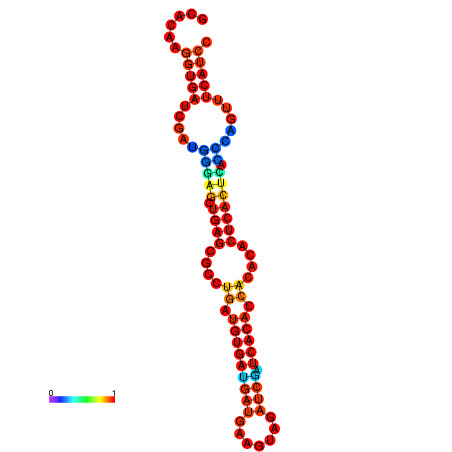
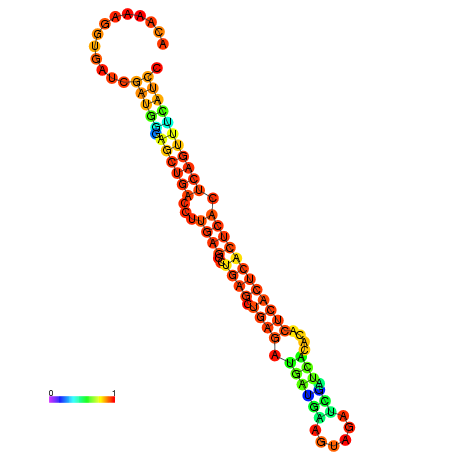
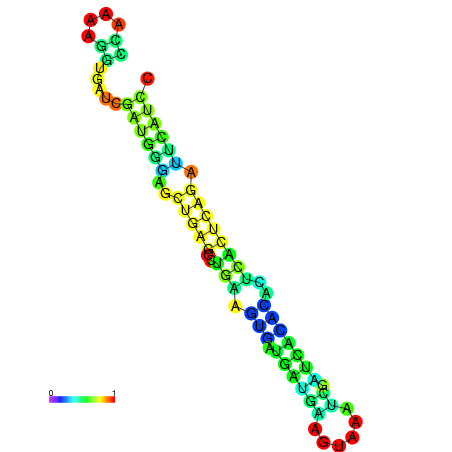
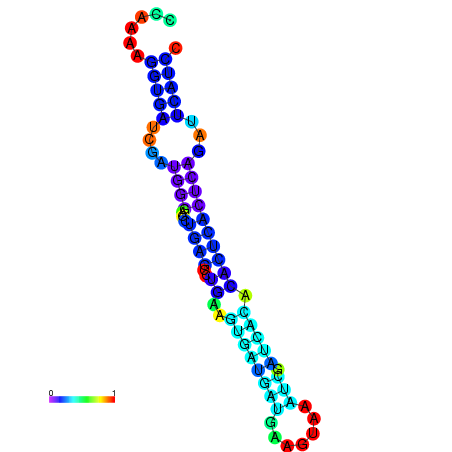
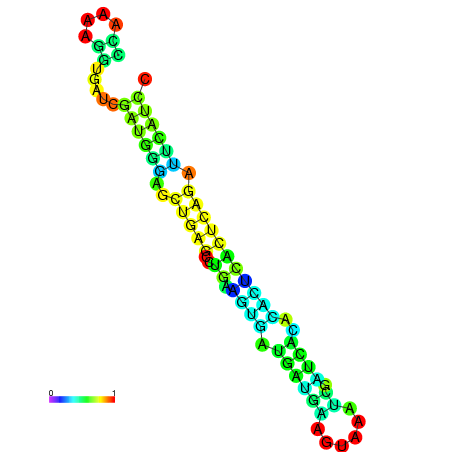
| dm3 |
chrX:16500125-16500242 + |
GTATTCGCAGTAC-AAGCACAAGGTGATCGATGGGAGATGA----GCGCCTGA--TGTGATGATGAAGTAGATCGATCACACCAC--ACACTCACTCA--------------------------CCCAGGTTTCATCCATACAAATGTGTTTG |
| droSim1 |
chrX_random:4339044-4339160 + |
GTATTCGCAATAC-AAGCACAAGGTGATCGATGGGAGCTGA----GCGCCTGA--TGTGATGATGAAGTAGATCGATCACACCAC--ACACTCACTCA--------------------------CCCA-GTTTCATCCATGCAAATGTGTTTG |
| droSec1 |
super_17:323461-323577 + |
GTATTCGCAATAC-AAGCACAAGGTGATCGATGGGAGCTGA----GCGCCTGA--TGTGATGATGAAGTAGATCGATCACACCAC--ACACTCACTCA--------------------------CCCA-GTTTCATCCATGCAAATGTGTTTG |
| droYak2 |
chrX:10606276-10606397 + |
GAATTTAGAATAA-AAACAAAAGGTGATCGATGGGAGCTGACCTTGAGCCTGAGCTGAGATGATGAAGTAGATCGATCACAC-AC--TCACTCACTCA--------------------------CTCA-GTTTCATCCATGCAAATTTGTTTG |
| droEre2 |
scaffold_4690:6858711-6858812 + |
CCC-----AATAA-AACCAAAAGGTGATCGATGGGAGCTGA----G--CCTGA--AGTGATGATGAAGTAAATCGATCACA-------CACTCACT------------------------------CA-GATTCATCCATGCAAATTTGTTTG |
| droAna3 |
scaffold_13337:17013621-17013746 + |
GAAATCTCAGCAGCAAGCTCAG-CGGATCG---GGTGTTGG----A--GGTGG--TCTGCCATTGACGAAGAGCAACAGCACCAA--TTCCTTCC---AGGAGCATAGCGCTCCCAAGC----------GTCGTCCCAGTATGGGTGCCTTGG |
| dp4 |
chrXL_group1a:8134511-8134553 - |
ACT------------------------------------------------------------------------------CCAC--ACACACACACA--------------------------CACA---GACAGACATATAAAGGA-CATG |
| droPer1 |
super_5:779941-779973 + |
ACACTCGCACTCC-C---ACAAGAAGAGCAGCAGTAG-------------------------------------------------------------------------------------------------------------------- |
| droWil1 |
scaffold_180698:6476640-6476654 + |
ATTT------------------------------------------------------------------------------------------------------------------------------------------GAATGTGTGTG |
| droVir3 |
scaffold_13042:4246856-4246902 - |
CAC-----------------------------------------------------------------------------AC-ACACACACACACATG--------------------------TACAGCTTTCACTCATGCATATTTGTT-- |
| droMoj3 |
scaffold_6500:26586811-26586907 + |
GCATTTGCACTG------------------CTCGAAAATGA----GCAATTGC--AAAGATCATTG-----------CACA---A--TACATTGC---AGAGAAATACTGCACAAAATAATTTAGTTA-ATTT------------TAAGTTGA |
| droGri2 |
scaffold_15245:5234538-5234618 - |
GCAAAATGAACAC-----AAAAACTGTTG------------------------------------------------CATACCAC--ACACTAACTG--T--AAATATCA--------------CATTTTTATTATTCTTATAATTATTAATA |