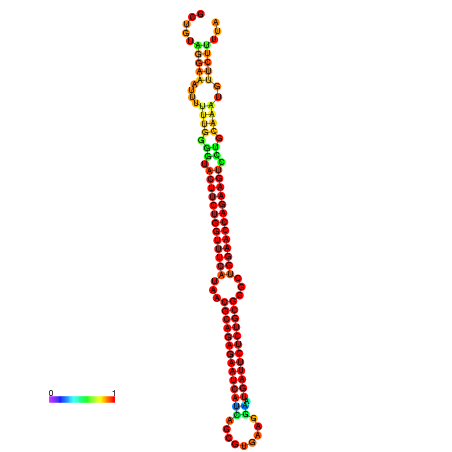
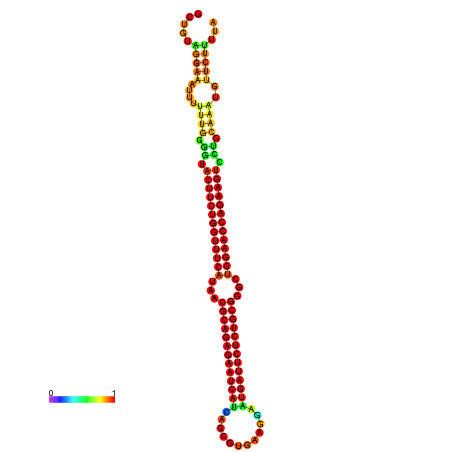
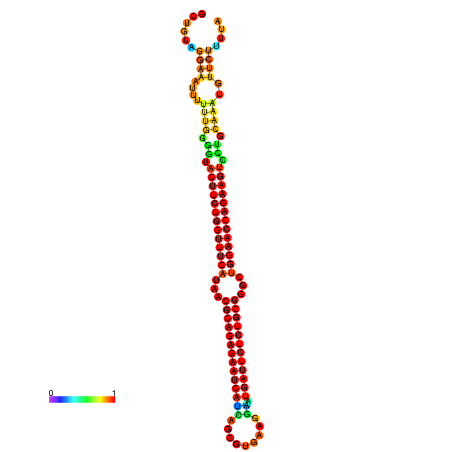
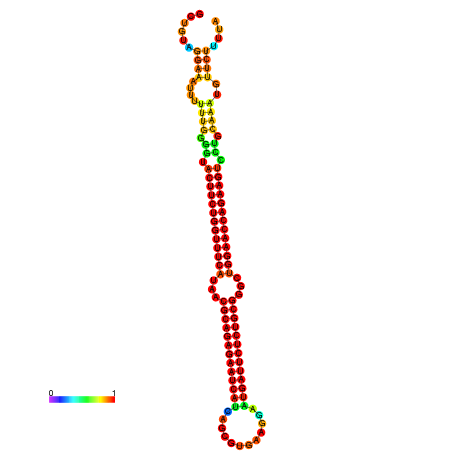
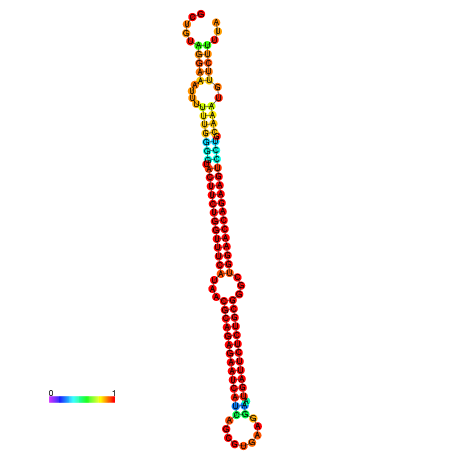
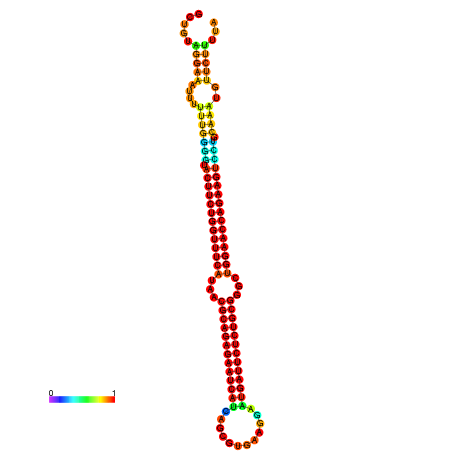
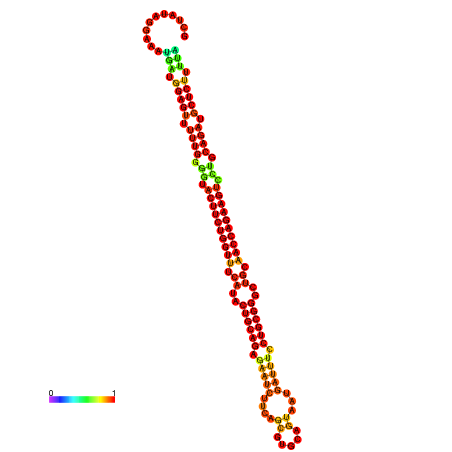
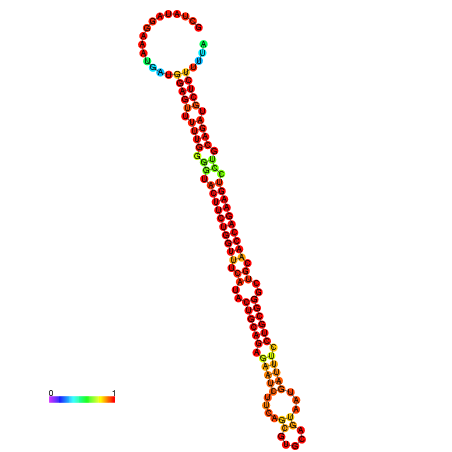
| dm3 |
chrX:5684770-5684909 + |
CACCCACTGCGAATTGCTGTAGGAAAT-------TTTTT-GGGGTACTTCTGGTTTCATAACGCAGAGAATCATC-----------AGCGTGAAGGAATGATTCTCTGCGGGCTGGAACCAGAAGTCCTGCAAATGTTCTTTTACGGATTTTAA---CCAGG |
| droSim1 |
chrX:4425250-4425396 + |
CACCCACTGCGAATTGCTATAGGAAATGATGGAGTTTTT-GGGGTACTTCTGGTTTCATACTGCAGAGAATCTTC-----------AGCGTGCAGTAATGATTTCCTGCGGGCTGCAACCAGAAGTCCTGCAGATGCTCTTTTACGGATTTTAA---CCAGG |
| droSec1 |
super_4:1079282-1079434 - |
CACCCACTGCGAATTGCTATAGGAAATGATGGAGTTTTT-GGGGTACTTCTGGTTTCATAACTCAGAGAATCTTCATGATTTCTTAATCA-----TAATGATTTCCTGCGGGCTGCAACCAGAAGTCCTGCAGATGCTCTTTTACGGATTTTAA---CCAGG |
| droYak2 |
chrX:2788911-2789034 - |
CACCCACTGCGAAATGCTATAGGACATGATGGAGCTTCTGGAGGTACTTCTGGTTTTATAATGCAGAGAATCGTA-----------ATCGTGAAGGAATGATTCTATGCGTACTGAAATCGGGAGTCCTGCAGAT--------------------------- |
| droEre2 |
scaffold_4690:3033374-3033523 + |
CACCCACTGCGAAATGCTATAGGAAACGATGGGGTTTCT-GGGGTACTTCTGGTTTTATAACGCACGGAATCTGA-----------ATCGTGAATGAATGATTCCTTGCGGACTGGAACCAGAAGTCCTGCAGAGGCTTCTTGAATGATTTGAAGTTTCGGA |
| droAna3 |
scaffold_13117:1117754-1117783 + |
CACCCACTGCGACGTACTTTAGAGAGTTGT------------------------------------------------------------------------------------------------------------------------------------ |
| dp4 |
chr3:13862643-13862665 + |
CACCAGCTGTGGGCTGCTTTAGG------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer1 |
super_4:1457976-1457998 - |
CACCAACTGTGGGCTGCTTTAGG------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 |
scaffold_181096:7345062-7345085 - |
AACCCACTGTGATGTCCTGTAAAA------------------------------------------------------------------------------------------------------------------------------------------ |
| droVir3 |
scaffold_13324:2924689-2924720 + |
CACGGCCTGTGGTGTGCTATAAATTTT------------------------------------------------------------------------------ATGTG---------------------------------------------------- |
| droMoj3 |
scaffold_6496:13805954-13805976 - |
CACCGACTGCGGCGTCCTCTGAG------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_15112:3786307-3786359 + |
CACCAGCTGCGGCGTGCTATAAATTC--------TTTTC-GTTGTATTTCTGGGTGTTCAAC---------------------------------------------------------------------------------------------------- |