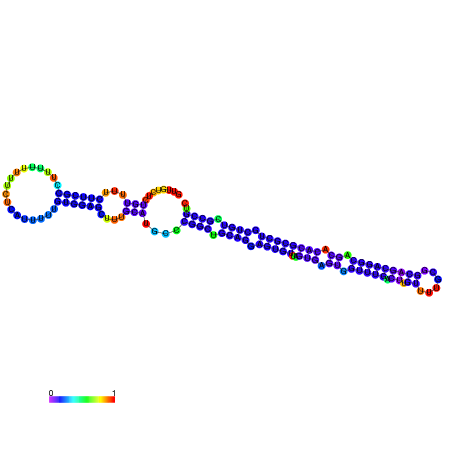
| dm3 |
chrX:3671399-3671553 + |
TGTGTTTGGTTGTTTTTCCTTTCTTTTC------------TCTCCGTCTCTCTCTTTC------------CCTCTCTCTGTT-TACATTGAGCTG-------TTTTGTCCGTGCAC----------GTGCTA---G---------------------ACGATGTAGAGAGTGAGG--GAGA---------------GAGCG---------------------AATAGAATAACAGCTCATTGTGCTGAG-CGCCCAGCC-T |
| droSim1 |
chrX:2692027-2692181 + |
TATGTTTGGTTGTTTTTCCTTTCTTTTC------------TCTCCGTCTCTCTCTTTC------------CCTCTCTCTGTT-TACATTGAGCTG-------TTTTGTCCGTGCAC----------GTGCTA---G---------------------ACGATGTAGAGAGTGAGG--GAGA---------------GAGCG---------------------AATACAATAACAGCTCATTGTGCTGAG-CGCCCAGCC-T |
| droSec1 |
super_13710:747-901 + |
TATGTTTGGTTGTTTTTCCTTTCTTTTC------------TTTCCGTCTCTCTCTTTC------------CCTCTCTCTGTT-TACATTGAGCTG-------TTTTGTCCGTGCAC----------GTGCTA---G---------------------ACGATGTAGAGAGTGAGG--GAGA---------------GAGCG---------------------AATACAATAACAGCTCATTGTGCTGAG-CGTCCAGCC-T |
| droYak2 |
chrX:4740636-4740793 - |
TGTGCTTGGTTGTTTTTCCTTTCTTTTC------------TCTCCGCCTCTCTCTTTC------------CCTCTCTCTGTT-TACATTAAGCGG-------TTTTGCCCGTGCAC----------GTGCTA---G---------------------ACGATATAGAAAGAGAGAGGGAGA---------------GAGCG--------------AA------TAACAATAACAGCTCGTTTTGCTGAG-CGCCCAGCC-T |
| droEre2 |
scaffold_4690:1061104-1061256 + |
TGTGTTTGGTTGTTTTTCCTTTCTTTTC------------TCTCCGTCT--CTCATTC------------CCTCTCTCTGTT-TACATTAAGCGG-------TTTTGCCTCTGCAC----------GTGCTA---G---------------------ACGGTATAGAGAGAGAGG--GAGA---------------GAGCG---------------------ATTACAATAACAGCTCATTGTGCTGAG-CGCCCAGCC-T |
| droAna3 |
scaffold_13337:5571517-5571673 - |
TGTGTTTTTGTTTTTGTTGTCTCTGTTT------------TCTTCGCCTTTTTTTTC-------------TC-ATTTTTG-------TGGAGCTTTGCATGCCCGGCTGCAGGAGT----------GTTAGT---GAGTGGTTTGACTTGTTTTGCGGCAGCAGGCAGCACACGC--GCTG---------------CTGT-------------------------------------CGCCGTCGCCGC-TGTCTAGC--- |
| dp4 |
Unknown_group_658:20580-20732 - |
TACATTTTTCCACTTGCCCACACTTTTCGCTGCGCT----GCTCTCTCGCTCTCTCTCTCTCTCATGCTCTC-------ACT-CTCAGAAAGCTG-------TGCAAT------------------GCACCATGCG---------------------ACGACTGAAAGCATGAGA--GTGA---------------G--CC--------------AG---CTTTCAGTGTGGCAGTTTTTTTCGTTG-------------- |
| droPer1 |
super_23:1560592-1560751 - |
GTTTCTCTGCCTCTCCCTCTC-------------------------TCCCTCTCTCTCTCTCTCGCTGTCTTCCTCTTTCTC-AACTATGTGTGG-------CCATGTGTGTGTGT----------GTGTGT---G---------------------TGTGGGTTGTATCTGTGT--GTGA---------------GAGCG-------------AAGCG---CAGACAATTAGT-TTTATTATGCTTTGTTGCCGAGCTTC |
| droWil1 |
scaffold_181096:11652056-11652210 + |
TCTGTTTGTGAGGGTCCCTCTCTTTTAC------------TCTCTGTCGCTCTCTCTC------------TCTCTCGCGCTG-CACAAAAAAATTAG-----TCATACC-AAAAACAAAATACAGATTGCAA---A---------------------GCAATAGAGAGAGAGAGG--GAGA---------------GAGAG---------------------AGA------GTAAGTGAGTGTG-AGAG-CGCCCA----- |
| droVir3 |
scaffold_12855:8711965-8712121 + |
TCTCTTTCTTTATTTCTCTGCTT--TTCTCTGTCTCTCTTTATCTGTATCTCTCTCTC------------TTTCTCTCTATTTTCTATTGCTCTG-------TTATGTC-AAACATTTGCTGCATACTTTCA---G---------------------GCGATATTAAAAGCGTGC--AAGA---------------TTTTC---------------------ATTTGTATAACTGCGTATT-------------------- |
| droMoj3 |
scaffold_6496:4311996-4312145 - |
TGTCTCTC--TATTTCTCTCTCTCTCTC------------TCTCTCTCTCTCTCTCTC------------TCTCTCTCTGCC-TGCCTTCA---C-------TTTTATACAAA---------------AATA---A---------------------TGGCCATCAAAATTAATA--AAAACGTAATCAGAGCGAATGACTACAGAGGAAAACGAAAAG--------TATAAAAG-------------------------C |
| droGri2 |
scaffold_15203:9842081-9842230 + |
TCCCTTCTCTCTCTC-------------------------TCTCTCTCGCTCTCTCTC------------TC------TCTC-TCTAT----TTG-------TCTCTTCCGCATA-----------GCGCAA---G---------------------TCGACGTCGGGGCTGACG--GGCA---------------GCGTTTCACAGGAGAATCGGGTGACTATCACCCTGATTGC-TGTTGTGCTG-C-TGTTCATCG-T |