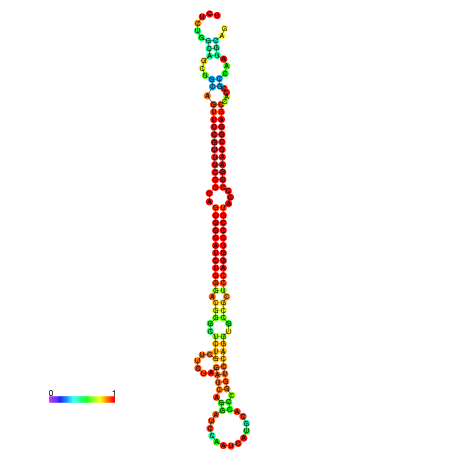
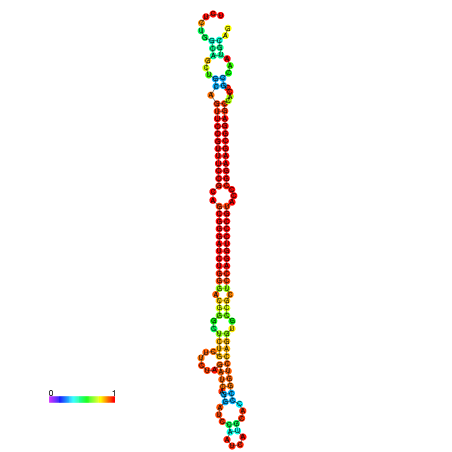
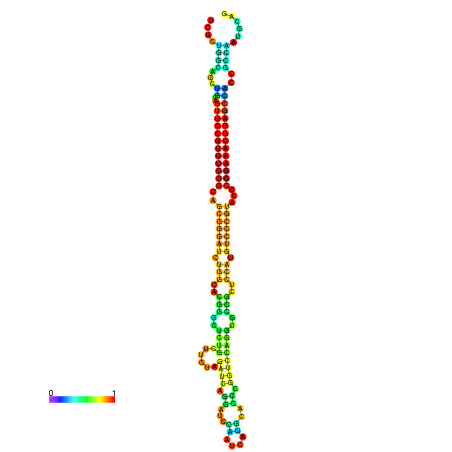
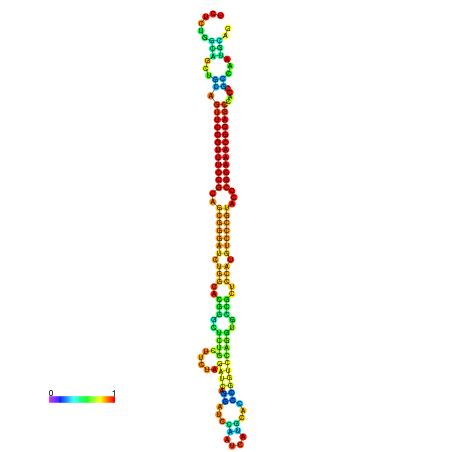
| dm3 |
chrX:15974940-15975104 - |
GTCCG------------------------------------A------ACCGTACGATCTCT--------------------------------GGCAGCTGCAGTTCCGTTT------CCGGAGCGGGATCTGGAACGGGCTCTGGTTCTAGATCAGGATCCAATCATGCACCCGGTCCAGGTACCGCTCCAGGTCCCGTTCCCGGAAACGGTGCCACCGCCAATGCAGCTGCAGCGGCATTCA |
| droSim1 |
wgm54c09.g1:392-557 + |
GTCCG------------------------------------A------ACCGCACGATCTCT--------------------------------GGCAGCTGCAGTTCCGTTT------CCGCAGCGGGATCTGGGACGGGCTCTGCTTCTAGATCAGGATCCAATCATGCACCCGGTCCAGGTGCCGCTCCAGGTCCCGTACCCGGAAGCGGAGCCACCGCCAATGCAGCTGCAGCGGCATTCA |
| droSec1 |
super_47:33133-33297 - |
GTCCG------------------------------------A------ACCGTACGATCTCT--------------------------------GGCAGCTGCAGTTCCGTTT------CCGCAGCGGGATCTGGCACGGGCTCTGCTTCTAGATCAGGATCCAATCATGCACCCGGTCCAGGTGCCGCTCCATGTCCCGTACCCGGAAACGGAGCCACCGCCAATGCAGCTGCAGCGGCATTCA |
| droYak2 |
chrX:10123735-10123899 - |
GTCCA------------------------------------A------ATCCCACGATCTCT--------------------------------GGCAGCTGCAGTTCCGTTT------CCGCAGCGGGTTCTGGAACGGGCTCTGGTTCTGGATCCGGATCCAATATGGCATCCGGTCCTGGTACCGCTCCAGTTCCCGTACCCGGAAACGGTGCCACCGCCAATGCAGCTGCAGCGGCCTTCA |
| droEre2 |
scaffold_4690:11103030-11103194 + |
GTCCA------------------------------------A------ATCCCACGATCTCT--------------------------------GGCAGCTGCAGTTCTGTTT------CCGGAACGGGTTCTGGAACGGGCTCTGGTTCGGGATCCGGATCCAATCTGGCACCCGGTCCTGGTACTGCTCCAGTTCCCGTACCCGGTAACGGTGGCACCGCCAATGCAGCTGCAGCGGCATTCA |
| droAna3 |
scaffold_13165:312527-312640 - |
GCATA------------------------------------A------GCCACATGGACTCT--------------------------------G---------------------------------------------------------GATCAGGCTCCGTATCTGCGTCAATTTTGGGCACCGGTCCAGGCCCTTTACATGGAAATAGTGCGACCTCGAATGTCTCTTCTGCACCCTTTA |
| dp4 |
chrXL_group1e:9575758-9575916 - |
GCCCC------------------------------------A------ATCCCAATGCCAGC-----------------------------AACGGCAGCTGTGGCTCCGCCTCTGCCTCCGGTTCGTCCAGTGGCTCG-GCACCGGCTCCGGCTCAGGGTCAGACTCAGGCCACGACTCAGGCACCG-----------ACTCACGGCAACACTTCGG---CCGGTGGCACAGCCTCGGCGTACT |
| droPer1 |
super_22:633193-633346 + |
GCCCC------------------------------------A------ATCCCAATGCCAGC-----------------------------AACGGCAGCTGTGGCTCCGCCT------CCGGTTCGTCCAGTGGCTCGGCACCGGTTCCGGGCTCAGGGTCAGACTCAGGCCACGACTCAGGCACC-----------AGCTCACGGCAACACTTCGG---CCGGTGGCACAACCTCGGCGTACT |
| droWil1 |
scaffold_181096:10624772-10624892 + |
CTGCATCATCGGCATCGCCAACTTATCAGAAGGGCAGTCCGAATCCAAATCCCAAT---------------------CTGAATGCAAATGCAACGGC--------------------------------------------------------------------------T------------------------------GGTGGCTCTGGAGCCAGTTCATCTGCTGCCACCGCTGCCTTTA |
| droVir3 |
scaffold_12472:523224-523310 + |
GCCCG------------------------------------A------ATCCCAATGGCGGT--------------------------------GGCTCCAGCGGTGCC------------------------------------------------------------------------------------AGTCTATCTGGCAGTAACAATGTGACTGCCACCGCTACAGCCGCCGCCTTTG |
| droMoj3 |
scaffold_6308:2944361-2944510 - |
GCCCG------------------------------------A------ATCCCAATGGCGGCGGCAGCGGCGCCGGTGG--------------------CGGTGGCGGTGCCGCTGCTGCTGGCTCCAACGGTGGC------------------------GGAAGTCT---------CTCGGGCAGCAGCAGCAGCAATGCAGCCACAGCCAATGCTACAGCCACCGCCGGTGGCGCGGGCTTTG |
| droGri2 |
scaffold_15203:4433131-4433217 - |
GCCCG------------------------------------A------ATCCCAATGGCTGT--------------------------------GTCTCCAGCGGTGCC------------------------------------------------------------------------------------AGTCTCTCTGGCAGCAACACTGTGACTGCCACCGCTACTGCCGCCGCCTTTG |