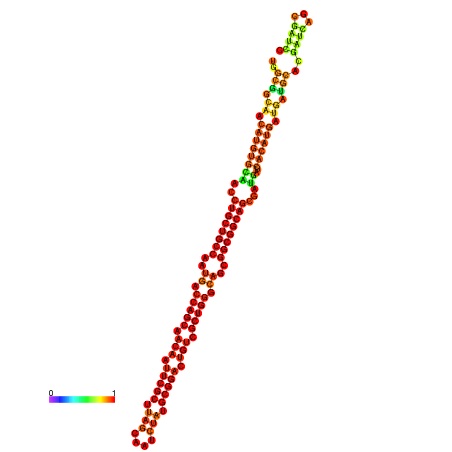
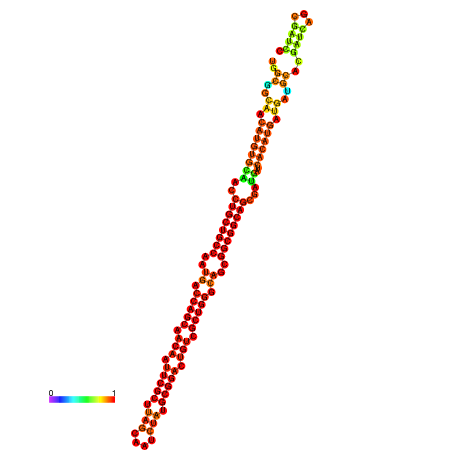
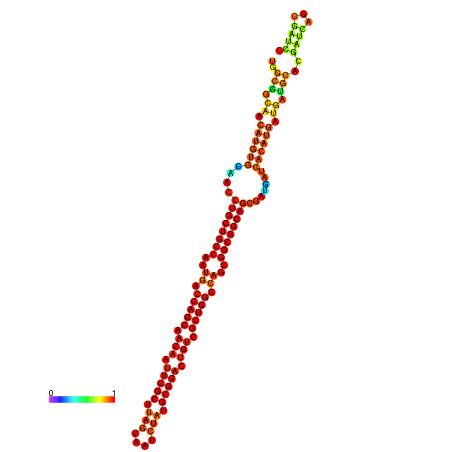
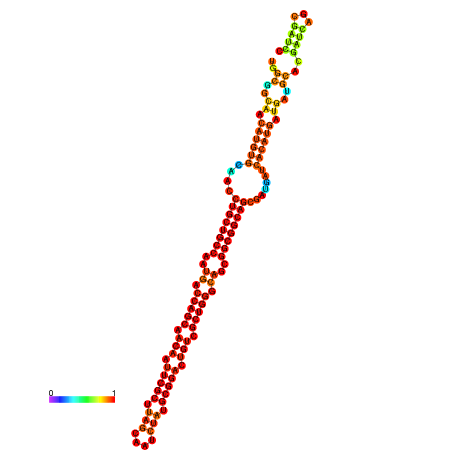
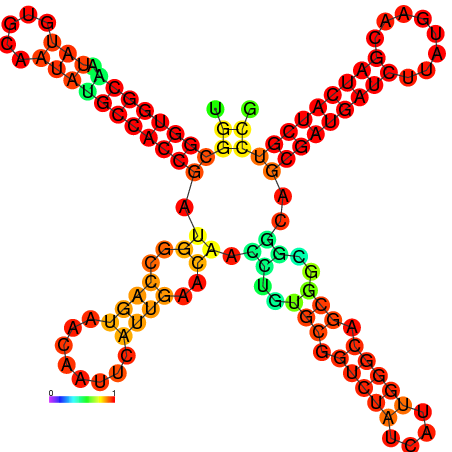
| dm3 |
chrX:6979754-6979901 - |
GAGTGCCAACAATTTAGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAACTCGCTGAATAATCTCTGCGGATTGTCGCTGGGCAGCGGTGGTAGTGATGATCTCATG------AACGATCCTCGGGCAAGCAACACCAACCTG |
| droSim1 |
chrX:5492772-5492919 - |
GAGTGCCAACAATCTAGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAACTCGCTGAATAACCTCTGCGGATTGTCGCTGGGCAGCGGTGGTAGTGATGATCTCATG------AACGATCCTCGGGCAAGCAACACCAACTTG |
| droSec1 |
super_24:199934-200081 - |
GAGTGCCAACAATCTAGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAACTCGCTGAATAATCTCTGCGGATTGTCGCTGGGCAGCGGTGGTAGCGATGATCTCATG------AACGATCCTCGGGCAAGCAACACCAACTTG |
| droYak2 |
chrX:14374793-14374940 + |
GAGTGCCAACAATCTGGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAATTCGCTGAATAACCTCTGCGGACTGTCGCTGGGCAGCGGCGGTAGCGATGATCTCATG------AACGATCCCCGGGCAAGCAACACCAACCTG |
| droEre2 |
scaffold_4690:15890244-15890391 - |
GAGTGCCAATAATCTTGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAATTCGCTGAATAACCTCTGCGGACTGTCGCTGGGCAGCGGCGGTAGCGATGATCTCATG------AACGATCCCCGGGCAAGCAACACCAACCTG |
| droAna3 |
scaffold_13117:2326296-2326455 - |
CAGTGCCAATTCCCTTGGCGGCGGCGGTGGCGGCAACATGTGCAACCTGCCGCCCATGACCAGCAACAATTCGCTTAACAATCTGTGCGGCCTGTCGCTGGGCAGCGGCGGCAGCGACGATCACATGCTT---AATGACCAGCGGCCGAGCAACACCAACCTG |
| dp4 |
chrXL_group1a:6391512-6391662 - |
CAGTGCCAATAATCTTG------------GCGGCAACATGTGCAACCTGCTGCCAATGACCAGCAACAATTCGCTTAGCAATCTATGCGGACTGTCGCTGGGCAGCGGCGGCAGCGATGATCACATGATGATGCACGATCAGCGATCGAGCAACACCAATCTG |
| droPer1 |
super_25:931278-931428 + |
CAGTGCCAATAATCTTG------------GCGGCAACATGTGCAACCTGCTGCCAATGACCAGCAACAATTCGCTTAGCAATCTATGCGGACTGTCGCTGGGCAGCGGCGGCAGCGATGATCACATGATGATGCACGATCAGCGATCCAGCAACACCAATCTG |
| droWil1 |
scaffold_180777:2984533-2984680 + |
CAGTGCCAATTCCCTTGGCG---------GTGGCAATATGTGCAATATGCCACCGATGGCCAGTAACAATTCATTGAACAACCTGTGCGGTCTATCATTGGGCAGCGGCGGCAGCGATGATCTTATG------AACGATCATCGTCCGAGTAATACCAATTTG |
| droVir3 |
scaffold_12928:60655-60799 - |
CAGCGCCAATTCCCTTG------------GCGGCAACATCTGCAATATGCCGCCAATGGCCAGCAACAATTCGCTGAACAATCTGTGCGGCCTATCGATTGGCAGCGGCGGCAGCGATGATCACATG------AACGATCAGCGCAACAGCAACACCAATTTG |
| droMoj3 |
scaffold_6359:1380681-1380818 - |
C-----CAATTCCCTCG------------GCGGCAACATCTGCAACATG---CCAATGGCCAGCAACAATTCGCTGAACAATCTGTGCGGCCTATCGATCGGCAGCGGCGGCAGCGATGATCTGATG------AACGATCAGCGCACAAGCAACACCAATTTG |
| droGri2 |
scaffold_14853:6692716-6692854 - |
CAGCGCCAATTCACTCGGCG---------G---A---CTCTGCAACATG---CCGATGGCCAGCAACAATTCGCTGAACAATCTGTGCGGCCTATCGATAGGCAGCGGGGGCAGCGATGATCTTATG------AACGATCCACGCAACAGCAACACCAATTTG |