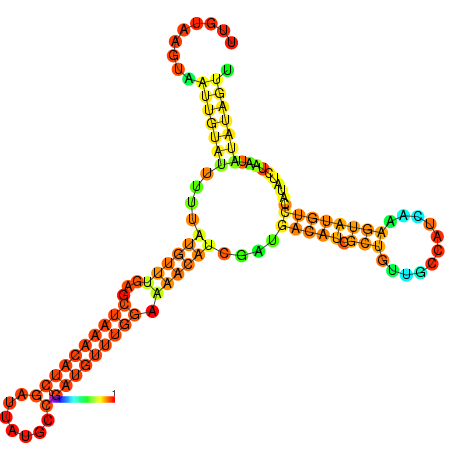
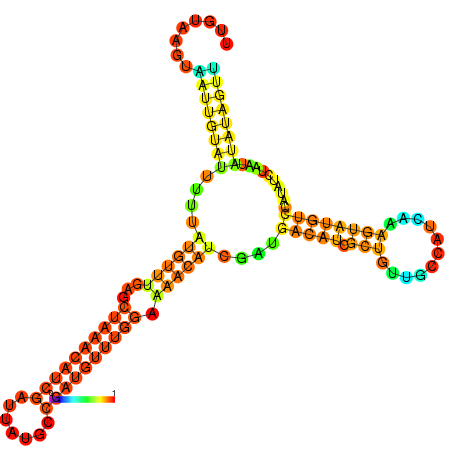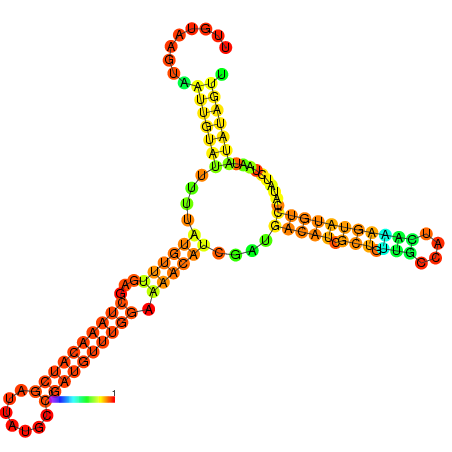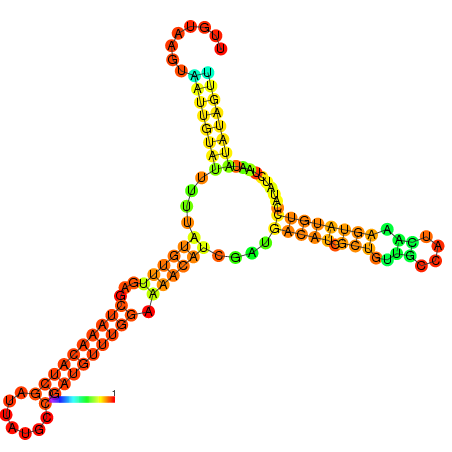| dm3 |
chr3L:23332312-23332452 + |
TATGACATCTGCCTGTT-----------ATGAA-------TACATATATT-TTTATGTTTG--AG-TCAACATC--GA------------------------------------------GTTGCCGATGTTTGG---AAAACATCGATGACATCGCT----------GTTGGCATCGA----AATATGTCCATATCTAATATATAGTTCCATTCTGTTTTATT |
| droSim1 |
chr3L:22456065-22456207 + |
AATGACAGCTGCCTGTT-----------ATAAG-------TAATTGTATT-TTTATGTTTG--AGCTAAACATC--GA-----------------------------------------TTATGCCGATGTTTGG---AAAACATCGATGACATCGCT----------GTTGCCATCAA----AGTATGTCCATATCTAATATATAGTTCCATTCTGTTATATT |
| droSec1 |
super_41:217199-217341 + |
AATGACATCTGCCTGTT-----------GTAAG-------TAATTGTATT-TTTATGTTTG--AGCTAAACATC--GA-----------------------------------------TTATGCCGATGTTTGG---AAAACATCGATGACATCGCT----------GTTGCCATCAA----AGTATGTCCATATCTAATATATAGTTCCATTCTGTTATATT |
| droYak2 |
chr3L:23917805-23917956 + |
TATGATATCTGAATATTTAGTTTTTCTTTTGC---------ATATTTATT-TTTAATATTG--AGCTTCACATA--TA-----------------------------------------TATTGCCGATGTTTAC---AAAACATCGATGACATTGTT----------TTTGGCATCGT----CATATGCCCATCACTAATATATAGTCCCCTTCTGTTTCATT |
| droEre2 |
scaffold_4784:22817455-22817604 - |
GGC---ATCTGTATAGG-----------ATGAA-------TAAATTAATT-TTTAAGTTTG--ACCACAACTTT--CA-----------------------------------------TTTTGCCGATGTTTAG---AAAACATCGATGACACCGCA-----TCATCGTTGTCATCGACTTACATATGTACATCTCTAATATATAGTATCATTCTGTTTTATT |
| droAna3 |
scaffold_12916:14167403-14167537 + |
TGCT---------CGTT-----------AGAGA-------TGAT-AAACA-TCGATGTTGGCTTGCCAAACATC--GA-----------------------------------------TGTCAACGATGTTTTC---AAAACATCGATGACATCGAT----------GTTAACATCGA----TGTTTCTCCACCTCTGACCACTGGAGTGAATTATTTTTAGT |
| dp4 |
chrXL_group1e:10317112-10317245 + |
TCAAACATATTATTTTT--------ATTGTAAATATCCTA----------------GTTAT--GGTGTAAATCT--GA-----------------------------------------AAATATCGATGTTTGAGAAAAAACATCGATGACATCCAT----------GTTAACATCGA----TGTTTGTCCAGCTTTAGCTTGCATCCCTAGCTTG------- |
| droPer1 |
super_69:203478-203603 - |
CATATT-----ATTATT-----------GTAAATATCCTA----------------TTTAT--AGTGTAAATCT--GA-----------------------------------------AAATATCGATGTTTGAGAAAAAACATCGATGACATCCAT----------GTTAACATCGA----TGTTTGTCCAGCTTTAGCTTGCATCCCTAGCTTG------- |
| droWil1 |
scaffold_181155:90645-90784 - |
TATTGTTTCCGCCTCTT-----------CTGAT-------CAATCTATTT-CTAATTTATT--TGCT-AACTT---CTTCTCGACTACGTACATATCTACTACGTACAACTCTAGTCAGATATATCCATATCCTATAACG-----------------------------------------------------------AAATCCAACTGCATTCTTAATCGTG |
| droVir3 |
scaffold_13036:829247-829384 - |
TTTATCGTTTATATTTGAAATT------ATTAA-------TTAAATTATT-ATTATTTTTA--AGTAATACAAA-----------------------------------------------------------------ATAAATTAAGGAAATAAAAGTGCAAATTTGTTGTTTTCAA----AG-ATACATTTAATTAATTTTTATTTTTTTTTTATTTTATT |
| droMoj3 |
scaffold_6500:19787300-19787437 - |
TATTATG---AGCTGAA-----------ATAAA-------TATAAAAATA-TTTA--ATTA--AATTCAACAAAATGATGTC------------------------------------------TAGATATTAAA---AAAAAAAAAAAAAATAAAAT----------ATATATATATA----TATATATATATATAT-ATATATATATATATTTTTTTTTTTT |
| droGri2 |
scaffold_14853:4224791-4224937 - |
TATATC-TATGCGTCTGTAGATACT----TGCA-------TAAGTGTATCATCTATGTACG--GTTTACATATA--GT-----------------------------------------GT---ATGGTTTATAA---AAAATATTTCTCAATACGCC----------GTTAGTACAAA----AAAAAAGTAGTGTATAAAAAATGCTTGCACTTTGTTTTAGT |