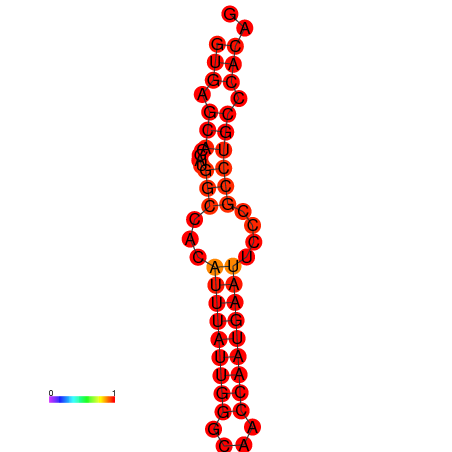
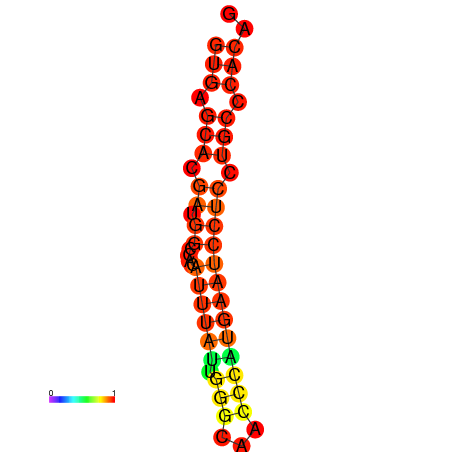
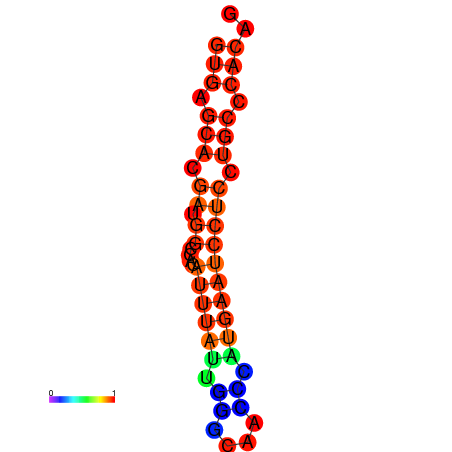
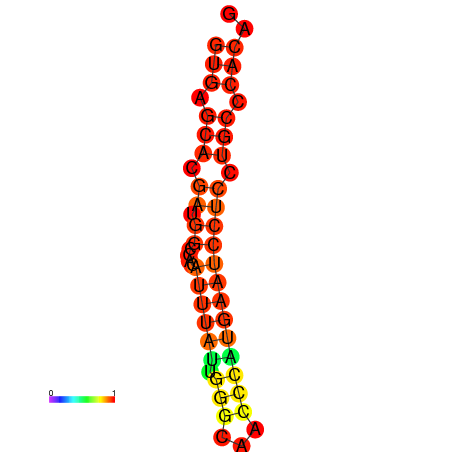
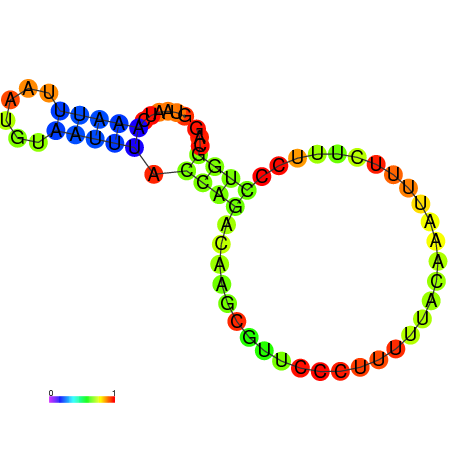
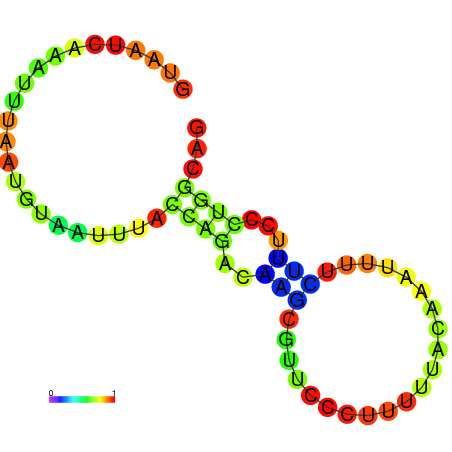
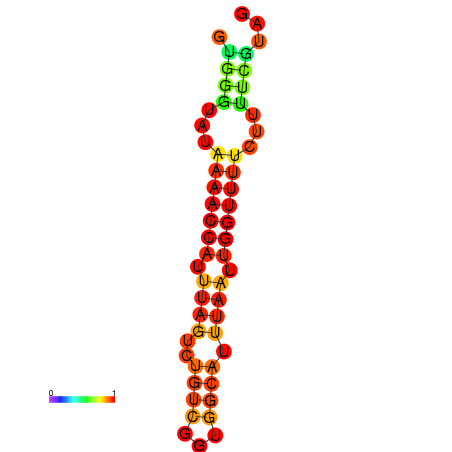
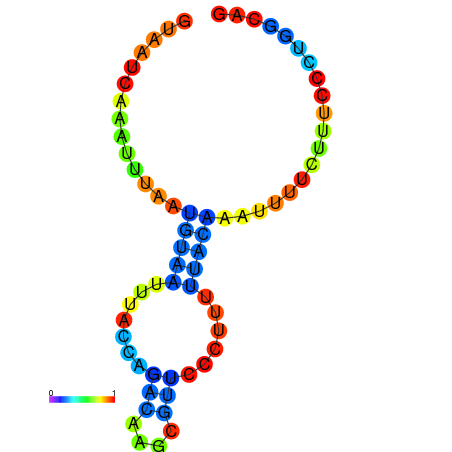| dm3 |
chr3R:4134138-4134222 - |
TCGCACATGTCTAAGGTGAG------CAC----GATGGCCACATTTA-----TTGGGCAA----------CCAATGAA-------TT--CCCGCCTGCC---CACAGGTTCACTCG--GTCTGC |
| droSim1 |
chr3R:17124439-17124521 + |
TCGCACATGTCCAAGGTGAG------CAC----GATGGCCACATTTA-----TTGGGCAA----------CCCATGAA-------TC--C--TCCTGCC---CACAGGTTCACTCG--GTCTGC |
| droSec1 |
super_0:17695137-17695219 + |
TCGCACATGTCCAAGGTGAG------CAC----GATGGCCACATTTA-----TTGGGCAA----------CCCATGAA-------TC--C--TCCTGCC---CACAGGTTCACTCG--GTCTGC |
| droYak2 |
chr3R:8209596-8209678 - |
GCGCACATGTCCAAGGTGAG------CAC----GGCGGCCCCA------AA--------ATCAAAGTTCTCTGACCAA-------A-----ATCCCGCC---CACAGGTTCACTCC--GTCTGC |
| droEre2 |
scaffold_4770:17439359-17439448 + |
GCGCACATGTCCAAGGTGAG------CAC----GATGGCCCCG------AC--------ATCGAAATT--C---TAAATCCCCAATCCAAAATCCCGCC---CACAGGTTCACTCA--GTCTGC |
| droAna3 |
scaffold_13340:13231716-13231803 - |
GCCCATCTGAACAAGGTAAT------TTG-AAT--TTGTGTTTT-TG-GCC-AGGAGCAA----------TTGATGTA-------TT--TGTTTATGTT---CGAAGGTACACTCT--GTTTGC |
| dp4 |
chr2:14657774-14657858 - |
GCCCACACGTCCAAGGTAAG------AA-----GCTAGTCGAA------AA--------ATTGAGGCATTTTCACGAA-------AC--A---ATTGTCTGAGGCAGGTTCACTCC--GTCTGC |
| droPer1 |
super_0:5758126-5758210 + |
GCCCACACGTCCAAGGTAAG------AA-----GCTGGTCGAA------AA--------ATTGAGGCATTTTCACGAA-------AC--A---AATGTCTGAGGCAGGTTCACTCC--GTCTGC |
| droWil1 |
scaffold_181089:2794443-2794538 + |
GCACATATGTCCAAGGTAAT------CAA-ATT--TAATGTAATTTA-----CCAGACAAGCGTTCCCTTTTTACAAA-------TT--TTCTTTCCCT---GGCAGGTTCACTCG--GTTTGC |
| droVir3 |
scaffold_12822:435887-435972 + |
CCACACATGTCAAAGGTGGG------TATAAAA--CCATTTAGT-CT-GTC-----------GGTGGCATTTAATTGG-------TT-----TTCTTTT---CGTAGGTGCATTCG--GTGTGC |
| droMoj3 |
scaffold_6540:27855886-27855971 - |
CAGCAA-TAGCAACAGCAAC------AGC----AATAGCAACAGCAACAGC----AGCAA----------CACCTGCAA------GC--CGCGGCAGCC---CAAAGTCTCGTTCG--CACCGC |
| droGri2 |
scaffold_15245:11304334-11304423 - |
ATGCACAGCTCCTTAAGCAATATGAACAG----CAGAGCCTGG------GTAGTGATGTA----------CC--TGGAT------CC--C-GGCCCATC---TACACGTTCAGTGGACATCAGC |