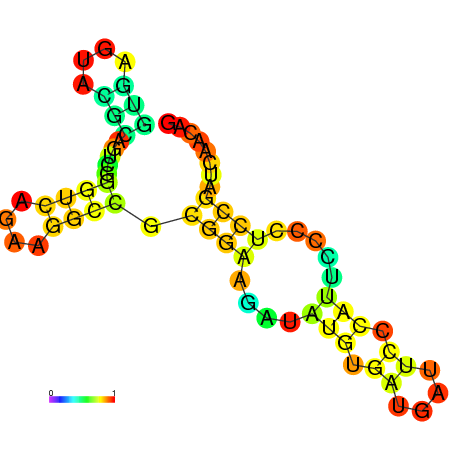
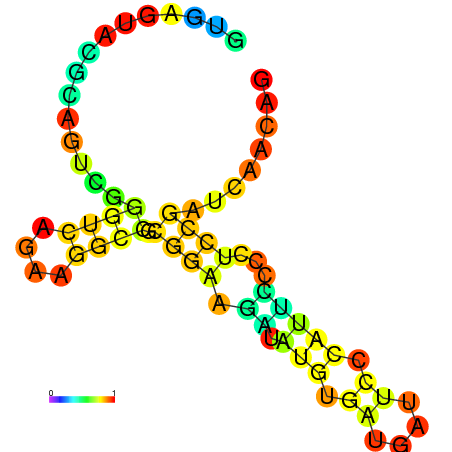
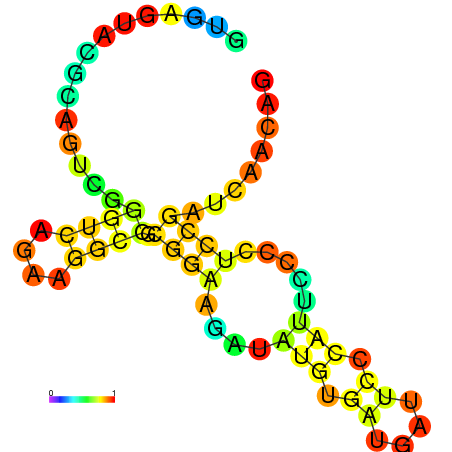
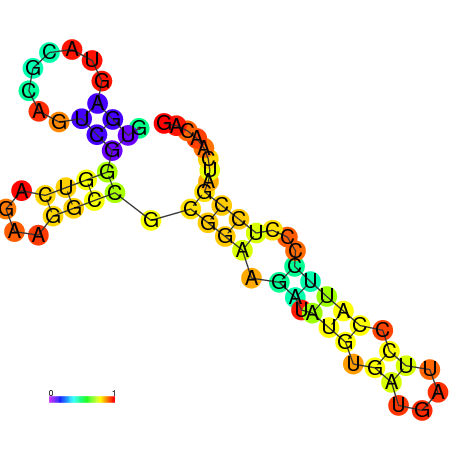
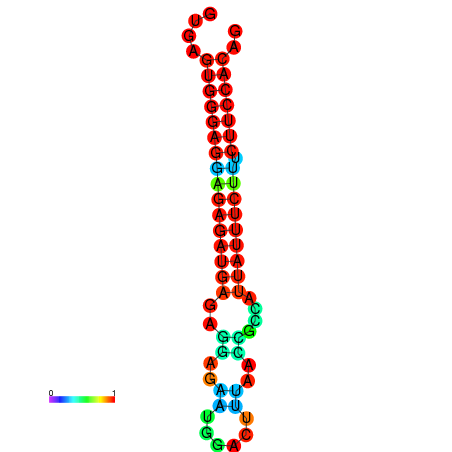
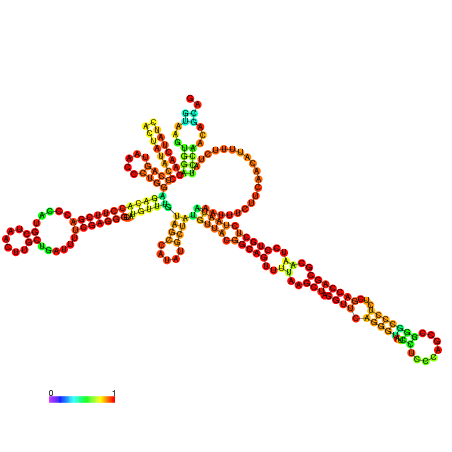
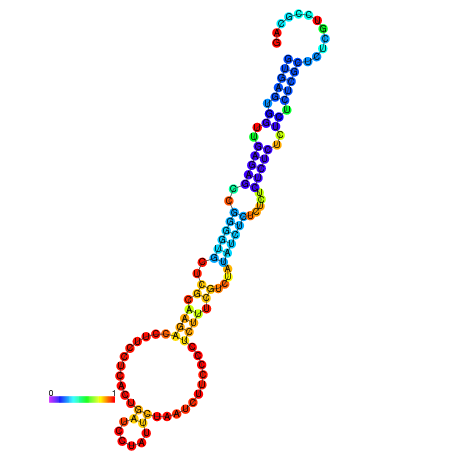| dm3 |
chrX:14093563-14093669 + |
GAGAGCCCCAGCAGAGTGAGTA-------GCGA---------------------------------------------------------------------------------TGGGG-GTCAGATGG-----------------------------------------------------------CCGTGGAA----GATATGTGACGATTGCC--------------------------------------------------------------------------------------------------CCCTTAC----------T---------------------ACTATCGTC---TGCTCAACAGGAAAGCAAACTACAT |
| droSim1 |
chrX_random:3764305-3764407 + |
GAGAGCCCC---AGAGTGAGTA-------GCGA---------------------------------------------------------------------------------TGGGG-GTCAGATG-------------------------------------------------------GATGGCCGTGGAT----GATATGTGACGATTGCC----------------------------------------------------------------------------------------------------CTTAC----------T------------------------ACGCTC---TGCCCAACAGGAAAGCAAGCTACAT |
| droSec1 |
super_20:697977-698075 + |
GAGAGCCCC---AGAGTGAGTA-------GCGA---------------------------------------------------------------------------------TGGGG-GTCAGATGG-----------------------------------------------------------CCGTGGAT----GATATGTGACGATTGCC----------------------------------------------------------------------------------------------------CTTAC----------T------------------------ACGCTC---TGCCCAACAGGAAAGCAAGCTACAT |
| droYak2 |
chrX:8347469-8347564 + |
GAGAGCCCC---AGAGTAAGTA-------GCCA---------------------------------------------------------------------------------GGGGG-ATCAGGAGG-----------------------------------------------------------CCGCGGAA----GGTATGTGATGATTCCC-----------------------------------------------------------------------------------------------------------------------------------------ATTACCCTC---CGATCCACAGGAAAGCAACCTACAT |
| droEre2 |
scaffold_4690:12836867-12836962 - |
GAGAGCCCC---AGAGTGAGTA-------CGCA---------------------------------------------------------------------------------GTCGG-GTCAGAAGG-----------------------------------------------------------CCGCGGAA----GATATGTGATGATTCCC-----------------------------------------------------------------------------------------------------------------------------------------ATTCCCCTC---CGATCAACAGGAAAGCAACCTACAC |
| droAna3 |
scaffold_13047:1616598-1616749 + |
GATACCCCG---AGAGTGAGTTAAATA-----GGTAGACTGGGTTTTACGGGTTTTGGGGATTAGGGTTAAGGGGTTTAAGGATCAGGGACTTGGCTA----------------GAAGG-ACCAA------------------------------------------------------------------AGGA-CTTA-----------------AGAA------------------------------------------------------------------------------------------------CTAAC----------A---------------------AAAATTCAC---CATTCCACAGGCAAGCACGGTACAC |
| dp4 |
chrXL_group1e:6144259-6144391 - |
GAAAACTCC---AGAGTGAGTTGAGTGAGGTT-----------------------------------------------------------------------------------GAGAGCCGGGGTG-------------------------------------------------------------CTCGGAA----GA----------CCATCCTC-------------------------------------------------------------------------ACTGATCCTATTCTAATCTT-------CCCCTCTTTCGTCTATATCTCTC--------------TCTCTC---TCGTCCGCAGGCAAGCGAACTATAC |
| droPer1 |
super_21:1109353-1109492 - |
GAAAACTCC---AGAGTGAGTG-------GTTG-----------------------------------------------------------------------------------AGAGCCGGGGTGC-----------------------------------------------------------TCGCAA------GA----------CCTTCCTC-------------------------------------------------------------------------ACTGATCCTATTCTAATCTT-------CCCCTCTTTCGTCTATATCTCTCTCTCTCTCTCTCTCTCGCTC---TCGTCCGCAGGCAAGCGAACTATAC |
| droWil1 |
scaffold_180702:2428501-2428590 + |
GAGAACACA---AGGGTGAGTG-------GGAG---------------------------------------------------------------------------------GAGAG-ATGAGAGGA-----------------------------------------------------------GAATGGA------------------------------------------------------------------------------------------------------------------------CTTTAAC-CGCCATTATT------------------------T--CTT---TCTTCCACAGGGAAGTGAACTACAA |
| droVir3 |
scaffold_12970:1543996-1544230 - |
GAAAACACA---CGAGTAAGTG-------GACAACTATC---------------------------ACTATACCCC-----------------------------------------------AG-TAACCCTGGAGACACCTTCGACCC-ATGCTAACTTGCTGATTTTCGAGGGCTATGTTTGTAG-----CC-ATAT-----------------GCTATGTTACGGCAGTTTTAAGCTAGGTTCAGG------GTA-TCCTCCCAGCCGGGCCCTCTCGACCAGCGCAATCCTGCTCTAATAAATTTC------TTCAA----------C------------------------ATTTTCTATCCAACAGCAGTCAAGCGAAATACAG |
| droMoj3 |
scaffold_6359:2739408-2739517 - |
GAAAACACA---AGAGTGAGTG-------ACAA---------------------------------------------------------------------------------GATTT-ATATATATT-----------------------------------------------------------TATAGGA-ATAT-----------------AATA------------------------------------------------------------------------------------------CTAATTCTTAT-CTACGTTATT------CCC------------GTCTCACTC---AATCAATTAGTCGAGCCGAATACAT |
| droGri2 |
scaffold_15081:1312788-1313066 - |
GAAAACAGA---GGAGTAAGTACGATC-----AGCTCGC---------------------------ATTATACCCCGTAAGAGAAGGTTCCCTAATGAAATGGACAAACAGAAATAGAA-ATCAATTT-----GAAAATCGGTGCAAGGTAAAATTAACTTGAT---CTCTCCAGCATAAATATATAT-----AT-ATAC-----------------GCTG------GACAG-ATTAAGTTCATTTTAGATGGTCGATAACACTTTCAACA--CCTTTTTAAATCAATTTCAAACTGCTTTTATTCAATTATT-------AACGTTTATCGTT------------------------T--TAT---CTGTCAACAGTCGAGCGAAATACAG |