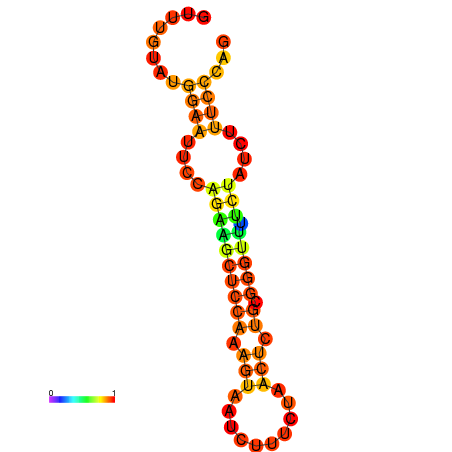
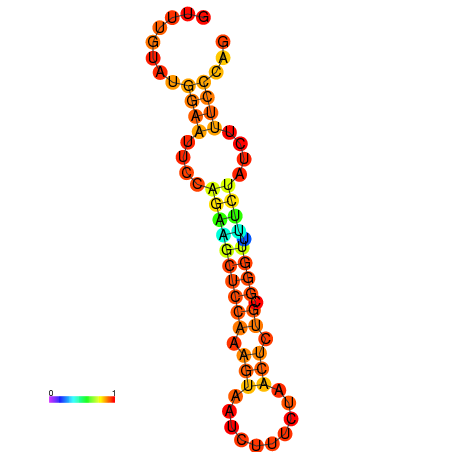
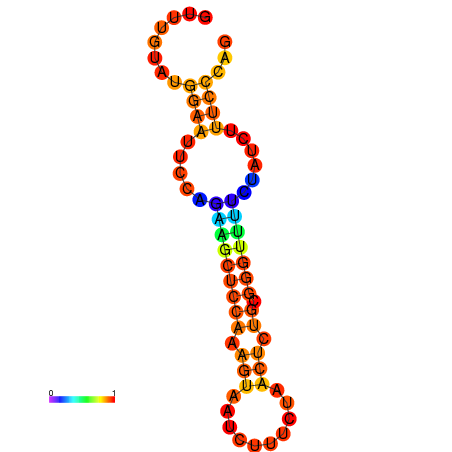
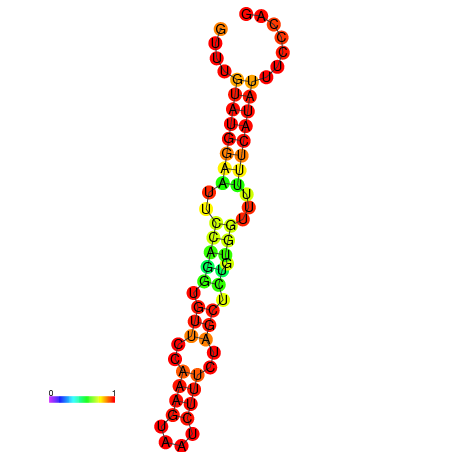
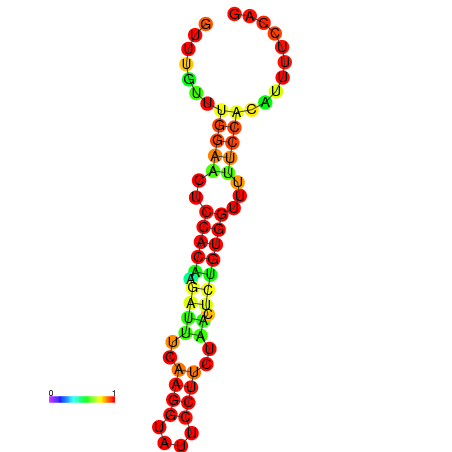
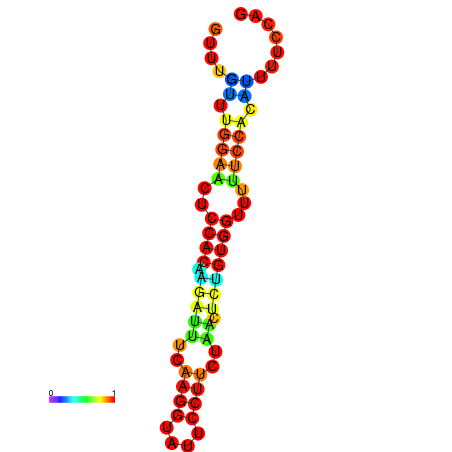
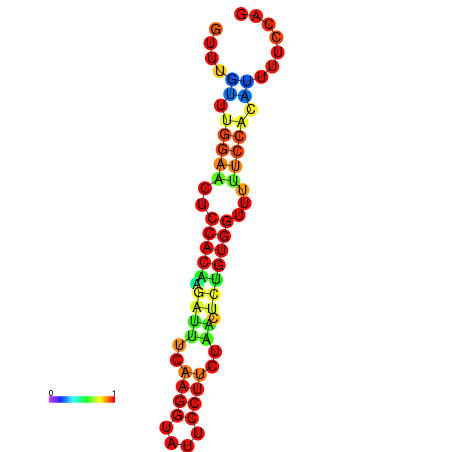
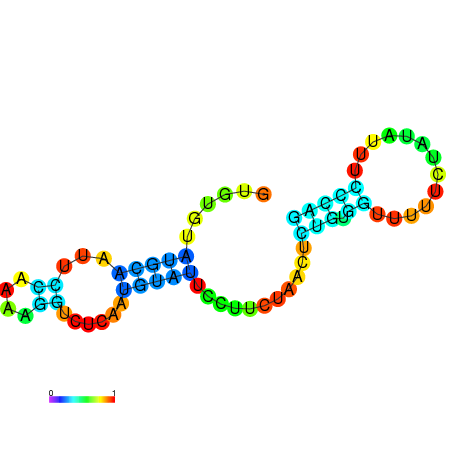
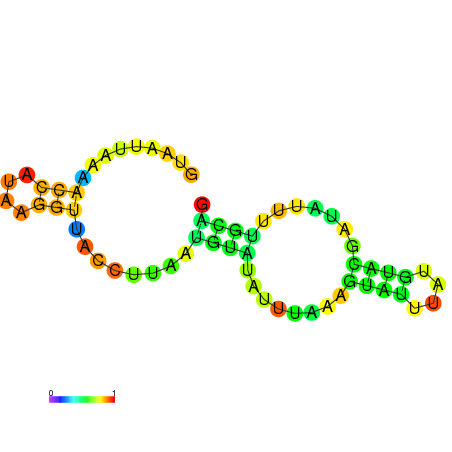
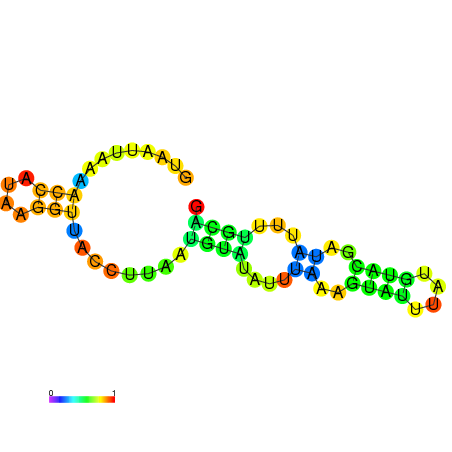
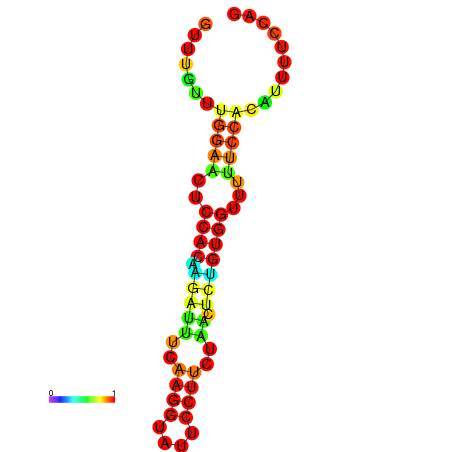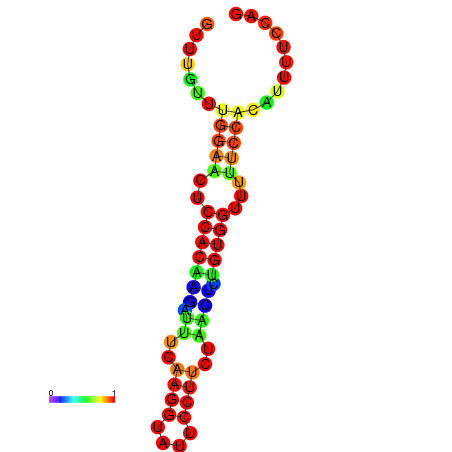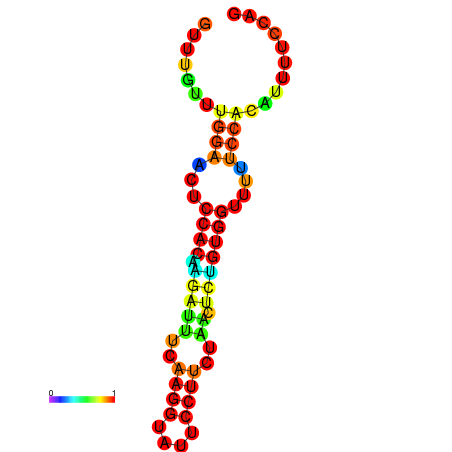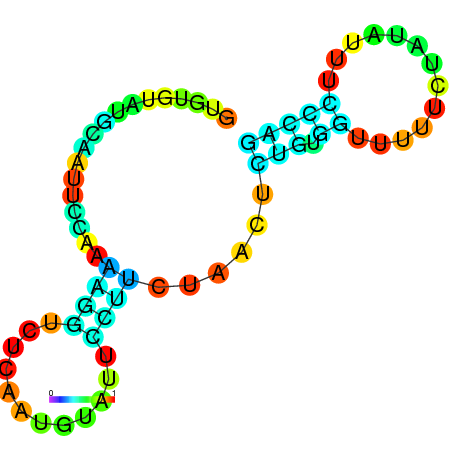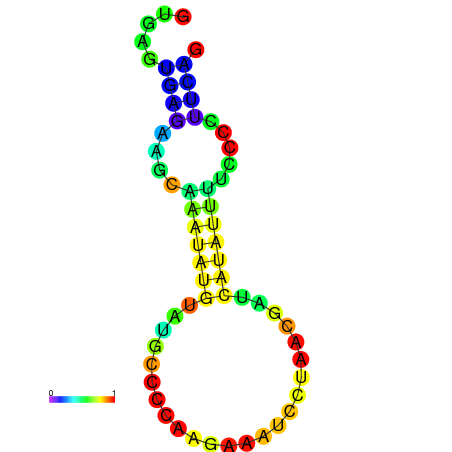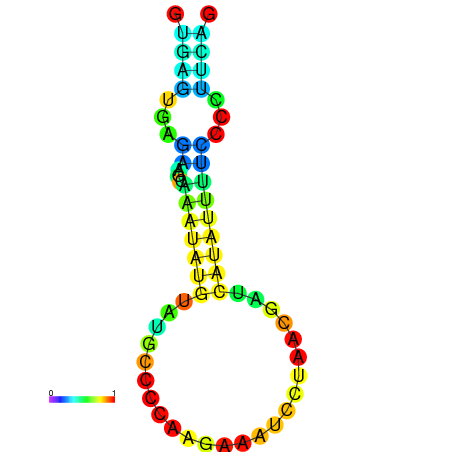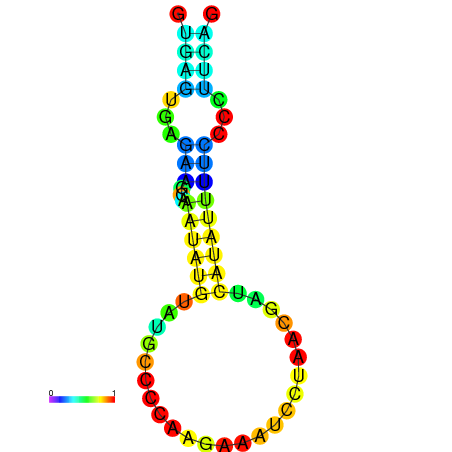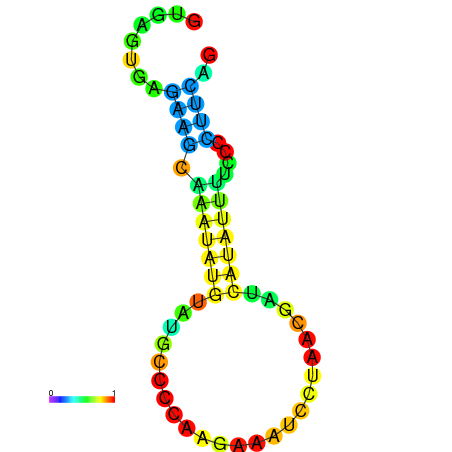| dm3 |
chr2L:4817801-4817896 - |
GGGTGAAACAGGCAGGTTTGTATGGAAT---TCCATGAGCTT--------------------------CAAAGTAATGCTTCTAACTCTGTGGTTTTTC-ATA-----TTTCCCAGTGGCAAGAGCACCCA |
| droSim1 |
chr2L:4687585-4687681 - |
GGGTGAAACAGGCAGGTTTGTATGGAAT---TCCAGAAGCTC--------------------------CAAAGTAATCTTTCTAACTCTGCGGGTTTTCTATC-----TTTCCCAGTGGCAAGAGCACCCA |
| droSec1 |
super_5:2913658-2913754 - |
GGGTGAAACAGGCAGGTTTGTATGGAAT---TCCAGGTGTTC--------------------------CAAAGTAATCTTTCTAGCTCTGTGGTTTTTTCATA-----TTTCCCAGTGGCAAGAGCACCCA |
| droYak2 |
chr2L:4827662-4827759 - |
GGGTGAAACAGGCAGGTTTGTTTGGAAC---TCCACAAGAT-------------------------TTCAAGGTATTCCTTCTAACTCTGTGGTTTTTCCACA-----TTTTCCAGTGGGAAAAGCACCCA |
| droEre2 |
scaffold_4929:4898740-4898837 - |
GGGTGAAACAGGCAGGTGTGTATGCAAT---TCCAAAAGGT-------------------------CTCAATGTATTCCTTCTAACTCTGTGGTTTTTCTATA-----TTTCCCAGTGGCAAGAGCACCCA |
| droAna3 |
scaffold_12916:9386286-9386375 - |
GGGCGAGACCGGCAGGTGAGAATACAAT---TCTACAACCCA--------------------------TACCCTTGTTTTCA------TATCATTTTTT-CAA-----TTTTTCAGTGGCAAGAGCACCCA |
| dp4 |
chr4_group3:3567830-3567920 - |
GGGCGAAACAGGAAGGTGAGTGAGAAGCAAAT----------------ATGTATGCCCCAAGAAATCCTAACGA----------------TC--------ATATTTTCCCCTTCAGCGGCAAGAGCACACA |
| droPer1 |
super_1:5048029-5048119 - |
GGGCGAAACAGGAAGGTGAGTGAGAAGCAAAT----------------ATGTATGCCCCAAGAAATCCTAACGA----------------TC--------ATATTTTCCCCTTCAGCGGCAAGAGCACACA |
| droWil1 |
scaffold_180772:7924021-7924113 + |
CGGAGAGACGGGCAGGTAATT----AAA---ACCATAAGGTTACCTTAATGTATATTTAAAGTATTTAT-----------------------G---TACGATA-----TTTTGCAGTGGAAAGAGCACTCA |
| droVir3 |
scaffold_13246:521378-521407 + |
GGGCGAGACGGGC-----------------------------------------------------------------------------------------------------AGCGGGAAGAGCACGCA |
| droMoj3 |
scaffold_6500:7585086-7585115 - |
GGGCGAGACGGGG-----------------------------------------------------------------------------------------------------TCTGGCAAAACGACGCA |
| droGri2 |
scaffold_15252:16729554-16729583 - |
GGGTGAAACTGGC-----------------------------------------------------------------------------------------------------AGTGGGAAGAGCACACA |