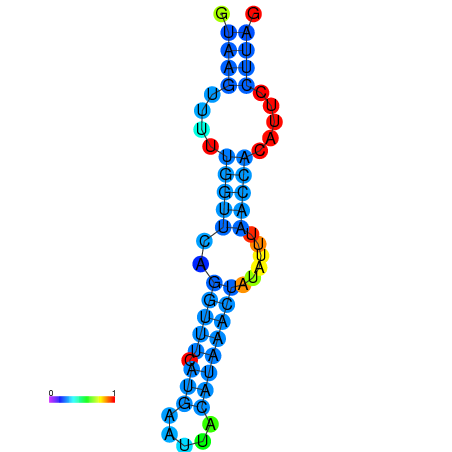
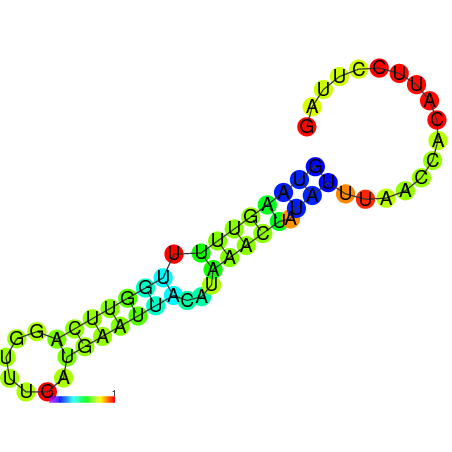
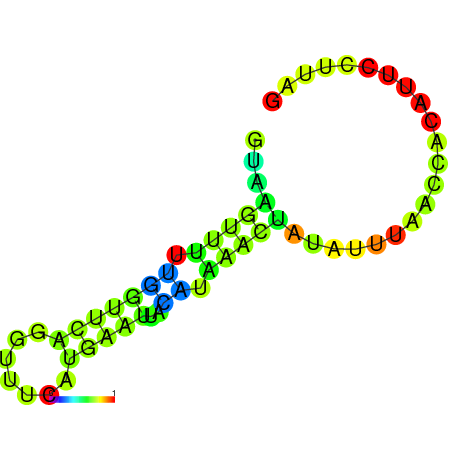
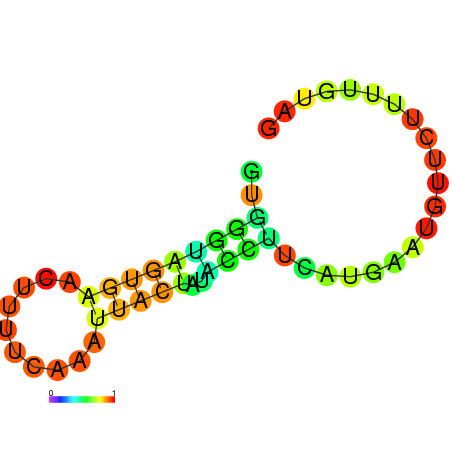
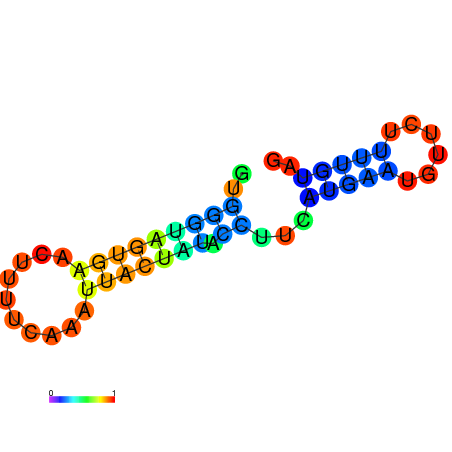
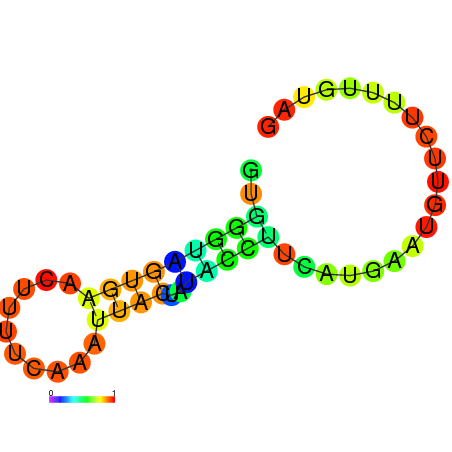
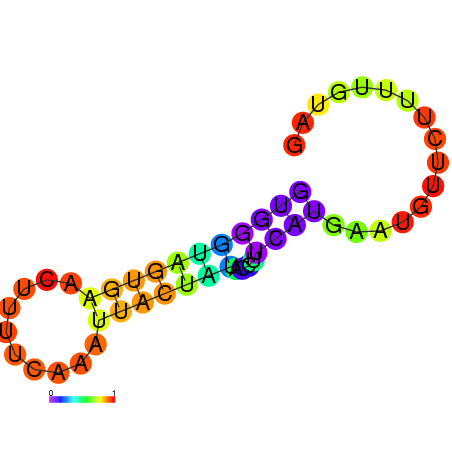
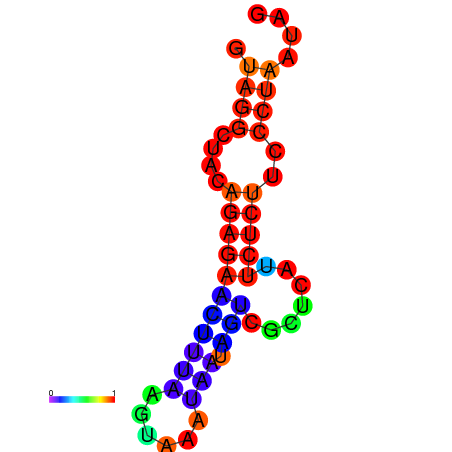
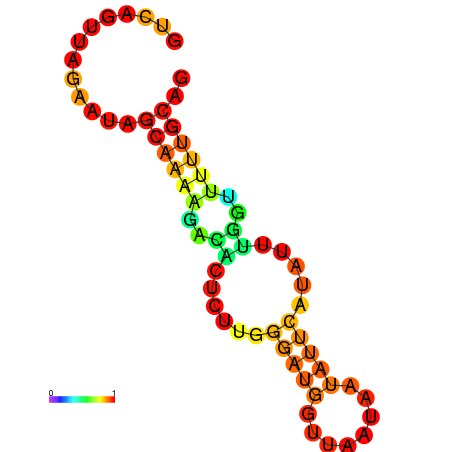
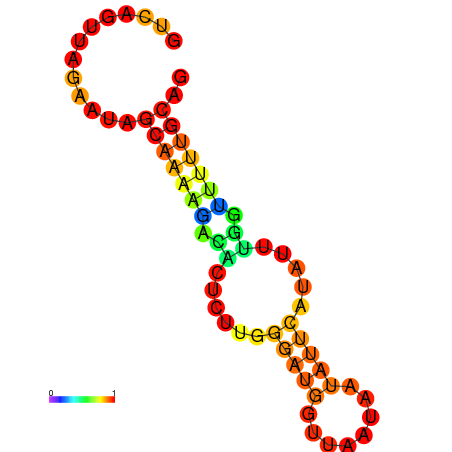
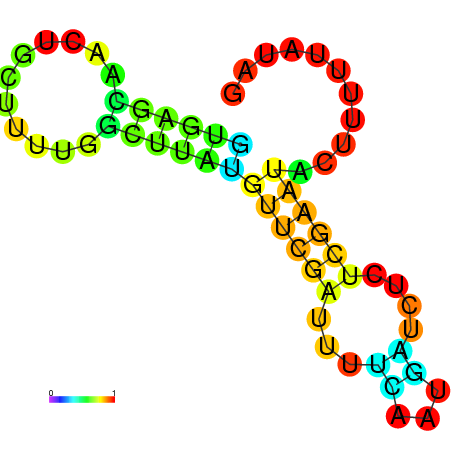
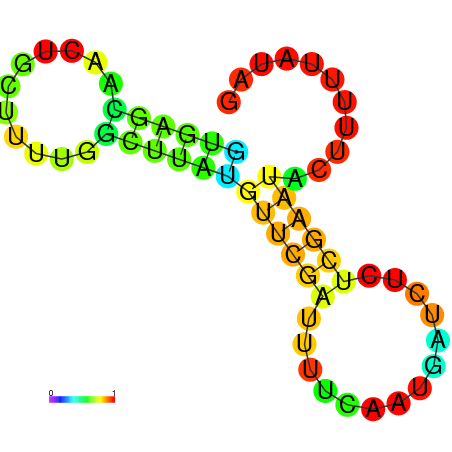
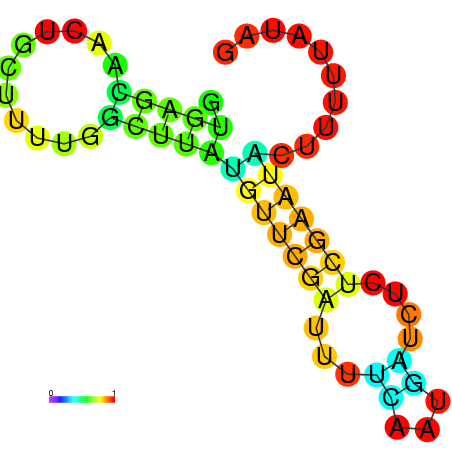
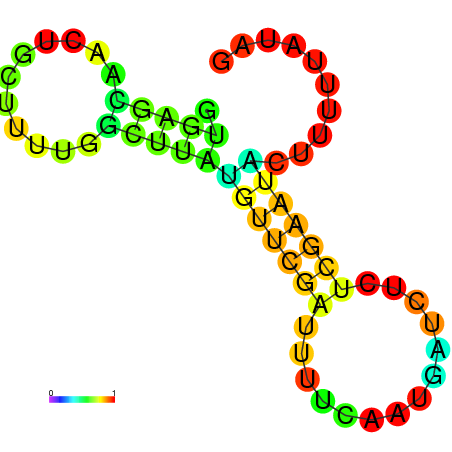
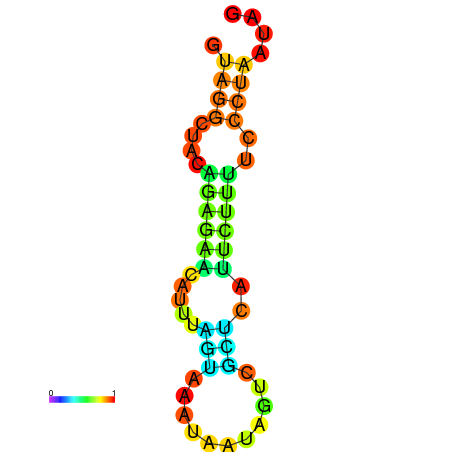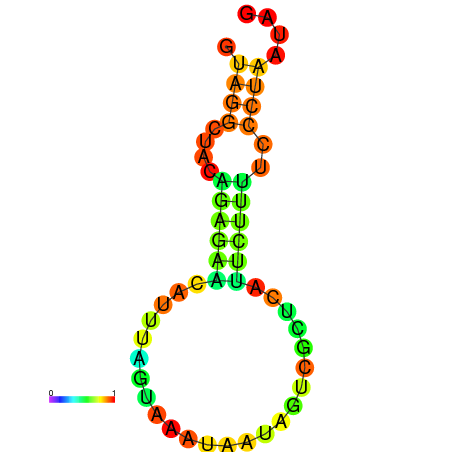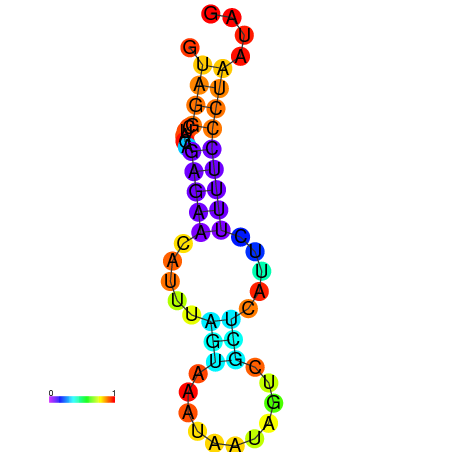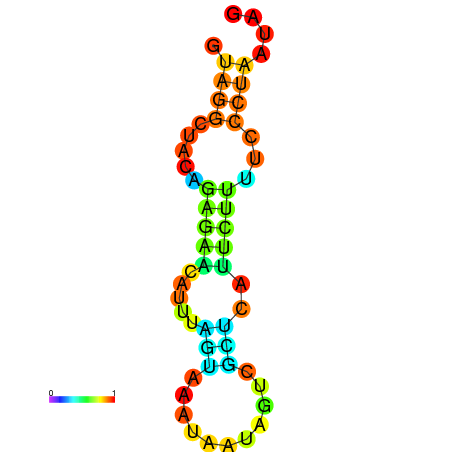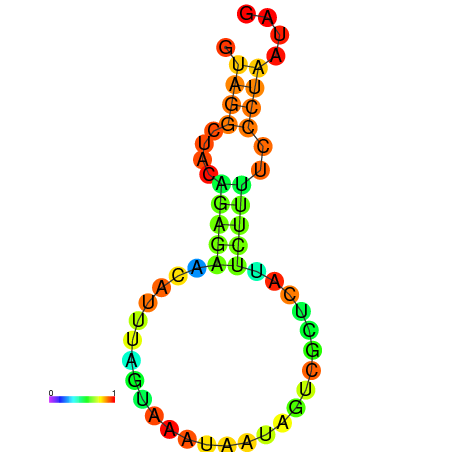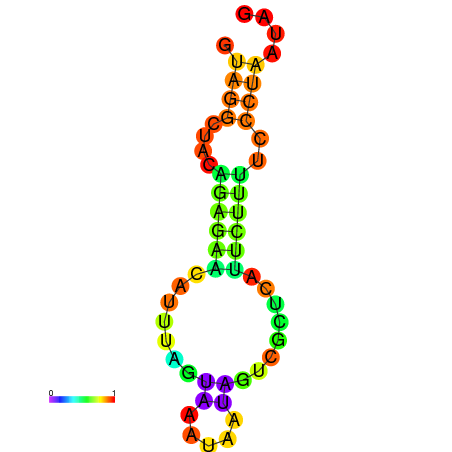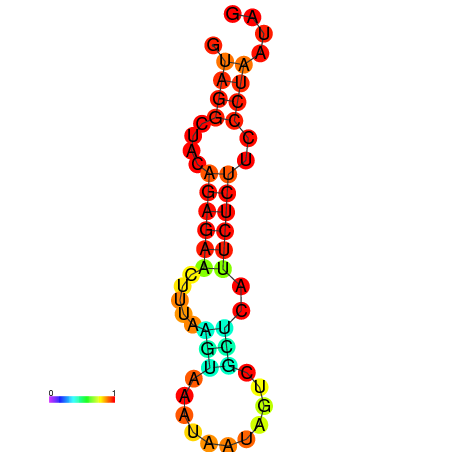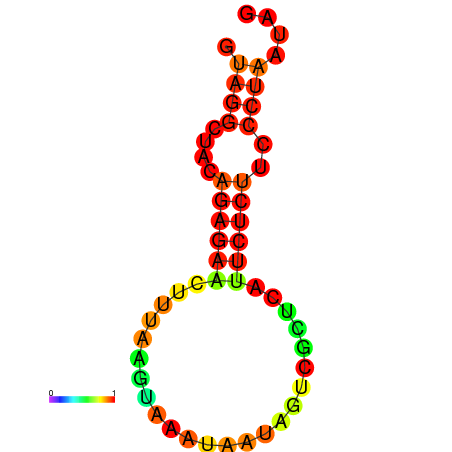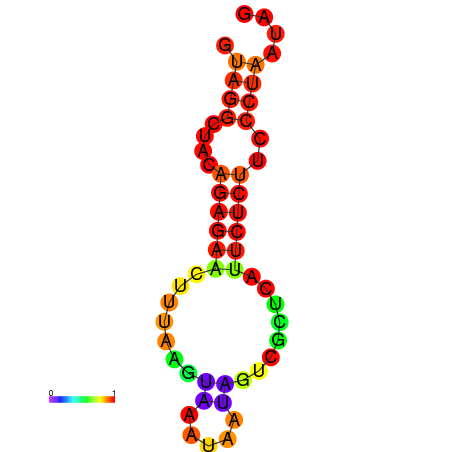| dm3 |
chr3R:11018004-11018094 - |
TTATTGCTGCAGGAGGTGAGTGATTAT----G-GTTTA----AGT-------AATACGAATTACATAAACTATATTTA-------ACCACTATTCTTAGATACAAAACACTGCT |
| droSim1 |
chr3R:10448012-10448102 + |
TTACTGCTGCAGGAGGTGAGTGTTTAT----G-GTTTA----AAT-------TATACGATTTACATAAACTATATTTA-------ACCACTACTCTTAGATCCAAAACACCGCT |
| droSec1 |
super_0:10933739-10933828 + |
TTACTGCTGCAGGAGGTGAGGTTTTAT----G-GT-TA----AGT-------TATACGATTTGCATAAACTATATTTA-------ACCACTACTCTTAGATCCAAAACACCGCT |
| droYak2 |
chr3R:15416636-15416724 + |
TTACTGCTTCAGGAGGTAAGTTTTT------G-GTTCA----GGT-------TTCATGAATTACATAAACTATATTTA-------ACCACATTCCTTAGATCCAAAATACCGCT |
| droEre2 |
scaffold_4770:10539530-10539618 - |
TTACTGCTTCAGGAGGTCAGTTTTT------G-GTTCG----ATT-------TCAACGATTTACATAAACTGTATTTA-------ACCATATTTCGTAGATCCAAAACACCGCT |
| droAna3 |
scaffold_13340:6107797-6107878 - |
TTGTTGCTGCAGGAGGTGGGTAGTG---------------AA---C------TTTTCAAATTACTATACC--------TTCATGAATGTTCTTTTGTAGATTCAAAATACTGCG |
| dp4 |
chr2:21002920-21003004 - |
CTGTTGCTGCAGGAGGTAGGCTACA------GAG------AA---C----ATTTAG---------TAAATAATAGTCGCTCATTC-TTTTCCCTAATAGATCCAAAACACTGCC |
| droPer1 |
super_3:3754054-3754138 - |
CTGTTGCTGCAGGAGGTAGGCTACA------GAG------AA---C----TTTAAG---------TAAATAATAGTCGCTCATTC-TCTTCCCTAATAGATCCAAAACACTGCC |
| droWil1 |
scaffold_181130:7245340-7245433 + |
TTGCTATTGCAAGAGGTCAGTTAGAAT----A-GCAAA----AGACACTCTTGGGATGGTTAATAATA----------TTCATAT-TTGGTTTTTGCAGATTCAAAATACTGCC |
| droVir3 |
scaffold_13047:7744104-7744193 - |
CTGCTGCTTCAAGAGGTGAGCAACTGCTTTTG-GCTTA----TGT----------TCGATTTTCAATGAT--------CTCTCG-AATACTTTTTATAGATACAAAACACAGCG |
| droMoj3 |
scaffold_6540:3682572-3682661 - |
TTGCTGCTGCAAGAGGTAAAATGTT------GAGCTCATAAAAGT----------TTCCTTT---TCAATTACA----TTCACAT-TCATTCGTTTTAGATACAAAATACGGCC |
| droGri2 |
scaffold_14830:3558248-3558343 - |
CTGCTGCTGCAGGAGGTGAGTTGCACCAACTG-ACACA----G-----TGTTTCCATAATTTACATTTTCTCTTTAT--------ATCTTTGTTCTCAGATACAAAACACGGCT |