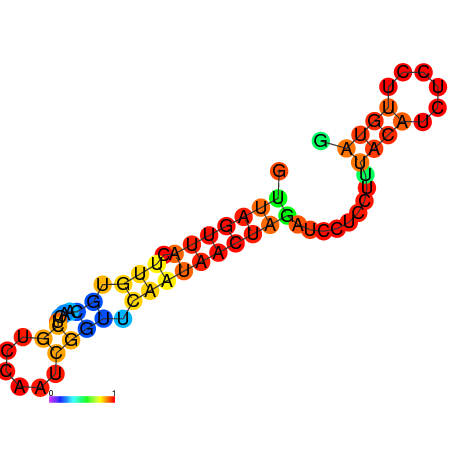
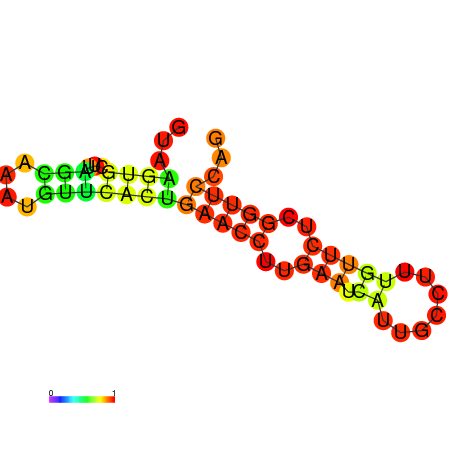
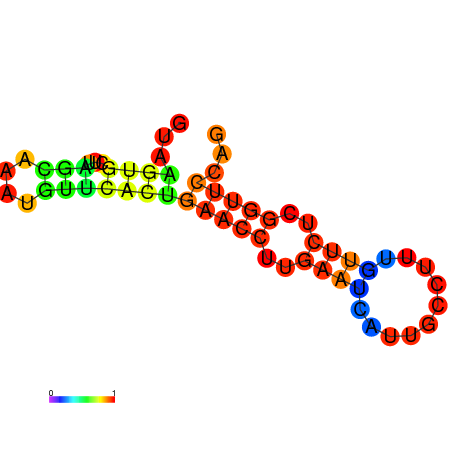
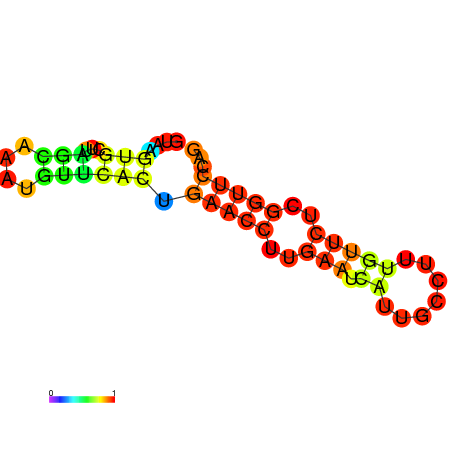
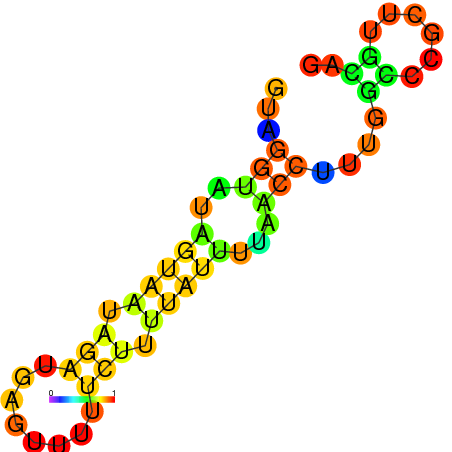
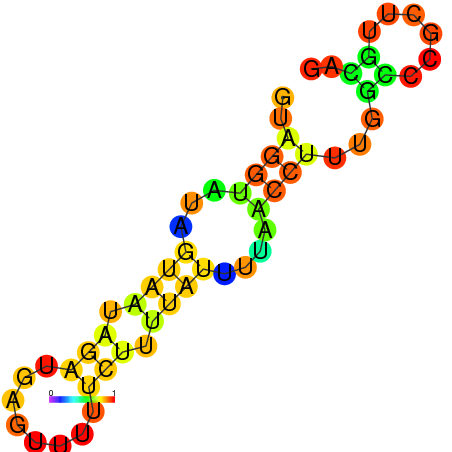
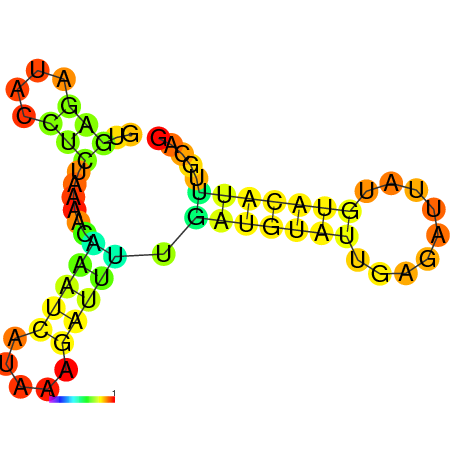
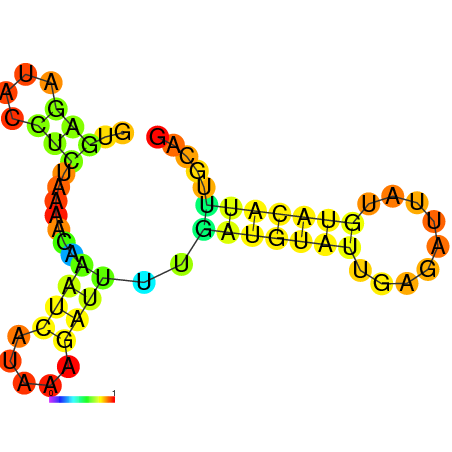
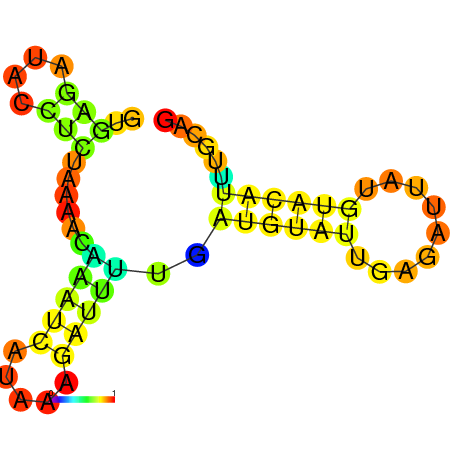
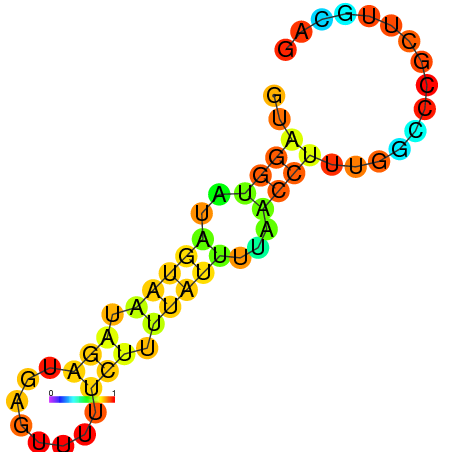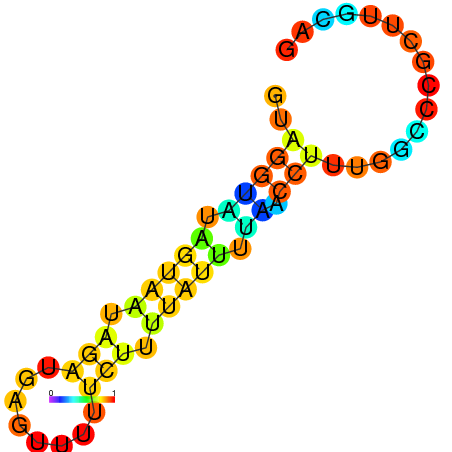| dm3 |
chr2R:13653883-13653980 - |
GAATCCAAGCTGAACGTGAGTTT-A--TCTAAATATG-CCCATTTATCAATAG-TCATAC----------TTATTTA-----T-------GC-TCCTTTA--ACTCCTTGCAGAGCTTCGAAAAAGAC |
| droSim1 |
chr2R:12393703-12393800 - |
GAGTCCAAGCTGAACGTGGGTTT-A--TCTAGATACG-CCCATTTATCATTGG-TCATAC----------TTATTTA-----T-------GT-TCCTTTA--ACTCCTTGCAGAGCTTCGAAAAAGAC |
| droSec1 |
super_1:11152590-11152687 - |
GAGTCCAAGCTGAACGTGGGTTT-A--TCTATTTACG-CCCATTTATCATTGG-TCATAC----------TTATTTA-----T-------GT-TCCTTTA--ACGCCTTGCAGAGCTTCGAAAAAGAC |
| droYak2 |
chr2R:15527673-15527770 + |
GAATCCAAGCTGAACGTGGGTTC-A--TCTAGATACT-TCCATATATCAAAAG-CCGTAC----------TTATTTA-----T-------GT-TATTTAA--ACTGCTTGCAGAGCTTCGAAAAAGAC |
| droEre2 |
scaffold_4845:7883019-7883116 - |
GAATCCAAGCTGAACGTGGGTTT-T--TCTAGATACG-CCCATTTATCAATGG-CCCTAC----------TTATTTA-----T-------GC-ATTTTAA--ACTCCTTGCAGAGCTTCGAAAAAGAC |
| droAna3 |
scaffold_13266:10531972-10532067 + |
GAATCCAAGCTGAACGTTAGTTACT--TGTGCA-A---CTCGT------------CCAATCGGTTC-AATAACTAGA-----T-------CC-TCCTTTACATCTCCTTGTAGGGCTTCGAGAAGGAC |
| dp4 |
chr3:16466931-16467019 - |
GAATCCAAGCTGAACGTAAGTGC-TTAGCAA---ATG-TTCACTGAACCTTGAAT----------------CATTGCCTTTG------------------TTCTCGGTTCCAGAGCTTCGAAAAGGAC |
| droPer1 |
super_4:295271-295359 + |
GAATCCAAGCTGAACGTAAGTGC-TTAGCAA---ATG-TTCACTGAACCTTGAAT----------------CATTGCCTTTG------------------TTCTCGGTTCCAGAGCTTCGAAAAGGAC |
| droWil1 |
scaffold_181141:4609865-4609965 + |
GAATCAAAATTGAATGTAAGCTG---------A-AAA------------TTTAATTT---ATATTTTGTTTTCTCCTATTTACATACCACGTGTCCTTCA--CTCAATTACAGAGCTTCGAAAAGGAT |
| droVir3 |
scaffold_12875:16919908-16919994 - |
GAATCCAAACTAAATGTGAGTAT-C--TCCTAA-TCGCCTCAT------------CCAAT----------TCATTTT-----T-------GA-CCCTTTG--CCTACTTTTAGAACTTTGAAAAAGAC |
| droMoj3 |
scaffold_6496:11084070-11084155 + |
GAATCTAAACTAAATGTAGGTAT-A--GTAA---TAG-ATGAGTTTTTCTTTTAT----------------TTTAACC-----------------TTTGG--CCCGCTTGCAGAACTTTGAGAAGGAC |
| droGri2 |
scaffold_15245:16459236-16459327 - |
GAATCCAAACTAAATGTGAGATACC--TCTAAA-ACAAATCAT------------AA---AGATTTTGATGTATTGAGATTA------------------TGTACATTTGCAGAACTTTGAGAAAGAC |