| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:18854056-18854169 + | GAGTTCCTTC------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTCATTG-TTTACC---AGTTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCCCTCAACGCTG----- |
| droSim1 | chr3L:18159364-18159477 + | GAGTTCCTTC------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTTATTG-TTTACC---AGTTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCTCTCATCGCTG----- |
| droSec1 | super_19:820227-820338 + | GAGTTCCTTC------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTTATTG---TACC---AGTTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCCCTCATCGCTG----- |
| droYak2 | chr3L:21787014-21787126 - | GAGTTCCG-C------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTTACTA-TTTAAC---AGCTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCTTACAACATTG----- |
| droEre2 | scaffold_4784:18670595-18670707 + | GAGTTCCTAC------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCT-ATTA-TCTAAC---AGCTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCTTACAACGCTG----- |
| droAna3 | scaffold_13337:18585473-18585582 - | GAGCTCCTAA------------ATGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTTATAA-ATTATT---AGCTGGCTTTCGAGCAATAATTGAAACCAAATAAGTGATCCTGACAAC--------- |
| dp4 | chrXR_group6:4103840-4103943 - | G---------------------CCGGACACTTATCTATTTTTGATTGTTGCTCAGAAAGCCCTTATAA-TTAACC---AGTTGGCTTTCGAGCAATTATCAAAGCCAAATAAGTGATCCCAACAACATT------ |
| droPer1 | super_9:2393048-2393151 - | A---------------------CCGGACACTTATCTATTTTTGATTGTTGCTCAGAAAGCCCTTATAA-TTAACC---AGTTGGCTTTCGAGCAATTATCAAAGCCAAATAAGTGATCCCAACAACATT------ |
| droWil1 | scaffold_180955:104826-104946 + | GGTTTGCTTATTTGCCCA---TCTGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTCATTACTTTATC---AGTTGGCTTTCGAGCAATAATTGAAACCAAATAAGTGAGCCTGTCAACA-------- |
| droVir3 | scaffold_13049:18083575-18083698 - | CATTTCCTTCCTACCTCACAAAACGGACGCCTATATCATTTTGATTGTTGCTCAGAAAGCCCTG--CA-TTCAAT---ACTTGGCTTTCGAGTAATAATCGAAACCAAATAAGCGATCCTAACTACGTTT----- |
| droMoj3 | scaffold_6680:23648550-23648660 - | ----------------------ACGGACCCTTATATAATTTTGATTGTTGCTCAGAAAGCCCGTAA---TTTAACTTAACATGGCTTTCGAGTAATAATCGAAACCAAATAAGGCATCCAAACATCATTTTGTCA |
| droGri2 | scaffold_15110:24135484-24135597 - | CAGCCAACTACT----------ATGGACGCTCATATAATTTTGATTGTTGCTCAGAAAGCCTTT--CA-CTTGAC---ACTTGGCTTTCGAGTAATAATCGAAACCAAATAAGCGATCCTAACTACAATT----- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:18854056-18854169 + | GAGTTCCTTC------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTCATTG-TTTACC---AGTTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCCCTCAACGCTG----- |
| droSim1 | chr3L:18159364-18159477 + | GAGTTCCTTC------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTTATTG-TTTACC---AGTTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCTCTCATCGCTG----- |
| droSec1 | super_19:820227-820338 + | GAGTTCCTTC------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTTATTG---TACC---AGTTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCCCTCATCGCTG----- |
| droYak2 | chr3L:21787014-21787126 - | GAGTTCCG-C------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTTACTA-TTTAAC---AGCTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCTTACAACATTG----- |
| droEre2 | scaffold_4784:18670595-18670707 + | GAGTTCCTAC------------ACGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCT-ATTA-TCTAAC---AGCTGGCTTTCGAGCAATAATTGAAACCAGATAAGTGATCCTTACAACGCTG----- |
| droAna3 | scaffold_13337:18585473-18585582 - | GAGCTCCTAA------------ATGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTTATAA-ATTATT---AGCTGGCTTTCGAGCAATAATTGAAACCAAATAAGTGATCCTGACAAC--------- |
| dp4 | chrXR_group6:4103840-4103943 - | G---------------------CCGGACACTTATCTATTTTTGATTGTTGCTCAGAAAGCCCTTATAA-TTAACC---AGTTGGCTTTCGAGCAATTATCAAAGCCAAATAAGTGATCCCAACAACATT------ |
| droPer1 | super_9:2393048-2393151 - | A---------------------CCGGACACTTATCTATTTTTGATTGTTGCTCAGAAAGCCCTTATAA-TTAACC---AGTTGGCTTTCGAGCAATTATCAAAGCCAAATAAGTGATCCCAACAACATT------ |
| droWil1 | scaffold_180955:104826-104946 + | GGTTTGCTTATTTGCCCA---TCTGGACACTTATATAATTTTGATTGTTGCTCAGAAAGCCCTCATTACTTTATC---AGTTGGCTTTCGAGCAATAATTGAAACCAAATAAGTGAGCCTGTCAACA-------- |
| droVir3 | scaffold_13049:18083575-18083698 - | CATTTCCTTCCTACCTCACAAAACGGACGCCTATATCATTTTGATTGTTGCTCAGAAAGCCCTG--CA-TTCAAT---ACTTGGCTTTCGAGTAATAATCGAAACCAAATAAGCGATCCTAACTACGTTT----- |
| droMoj3 | scaffold_6680:23648550-23648660 - | ----------------------ACGGACCCTTATATAATTTTGATTGTTGCTCAGAAAGCCCGTAA---TTTAACTTAACATGGCTTTCGAGTAATAATCGAAACCAAATAAGGCATCCAAACATCATTTTGTCA |
| droGri2 | scaffold_15110:24135484-24135597 - | CAGCCAACTACT----------ATGGACGCTCATATAATTTTGATTGTTGCTCAGAAAGCCTTT--CA-CTTGAC---ACTTGGCTTTCGAGTAATAATCGAAACCAAATAAGCGATCCTAACTACAATT----- |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| -34.2 |
|---|
 |
| -34.2 |
|---|
 |
| -33.5 |
|---|
 |
| -31.5 | -31.0 | -30.6 |
|---|---|---|
 |
 |
 |
| -31.6 | -30.9 | -30.8 |
|---|---|---|
 |
 |
 |
| -31.6 | -31.5 | -31.1 | -31.0 | -30.7 | -30.7 |
|---|---|---|---|---|---|
 |
 |
 |
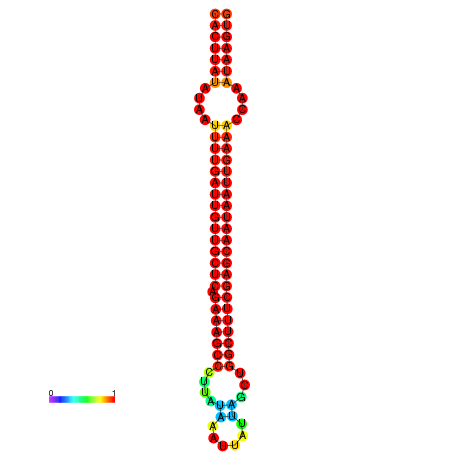 |
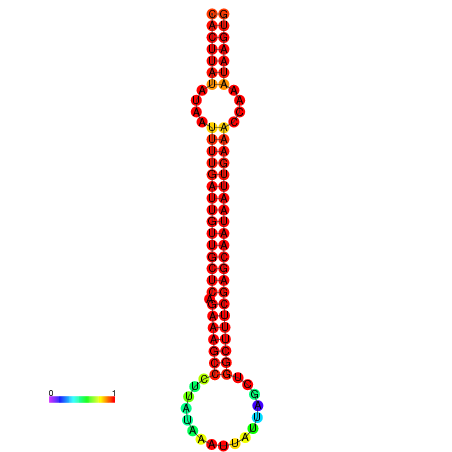 |
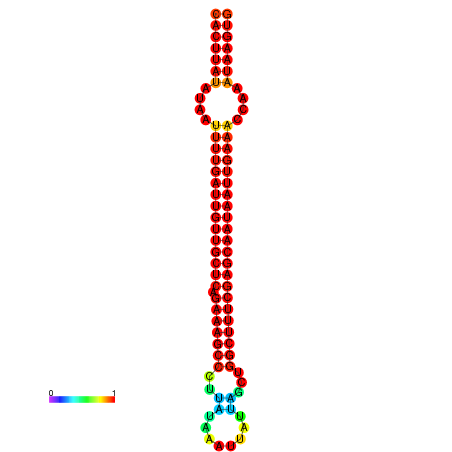 |
| -29.7 | -29.1 | -28.9 | -28.8 |
|---|---|---|---|
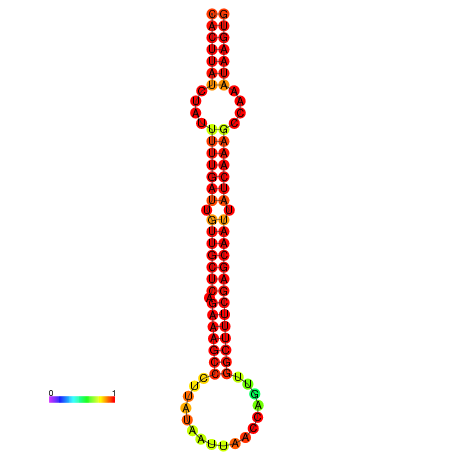 |
 |
 |
 |
| -29.7 | -29.1 | -28.9 | -28.8 |
|---|---|---|---|
 |
 |
 |
 |
| -31.0 | -30.8 | -30.7 |
|---|---|---|
 |
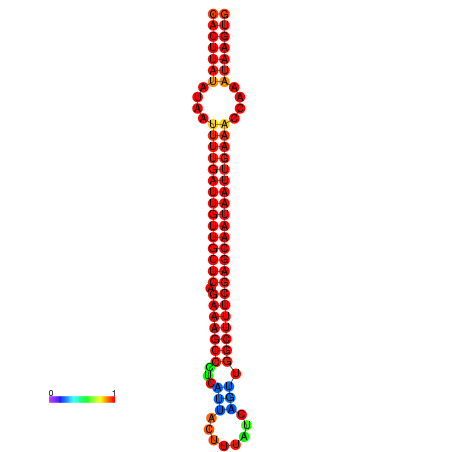 |
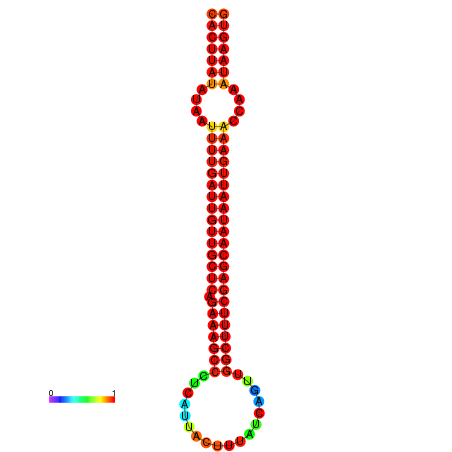 |
| -29.0 |
|---|
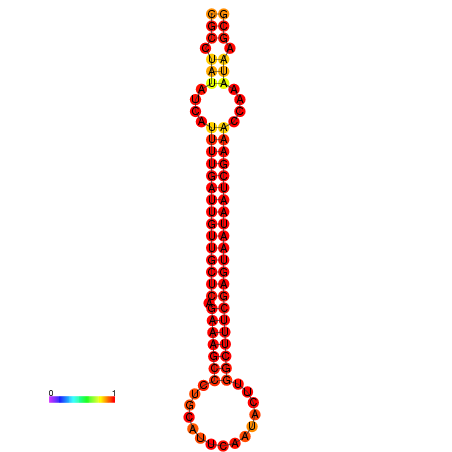 |
| -31.9 | -31.7 | -31.6 | -31.1 | -31.1 |
|---|---|---|---|---|
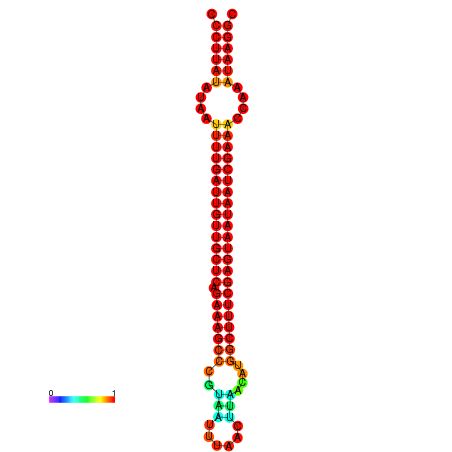 |
 |
 |
 |
 |
| -30.6 | -30.4 | -29.7 | -29.6 |
|---|---|---|---|
 |
 |
 |
 |
Generated: 01/05/2013 at 05:17 PM