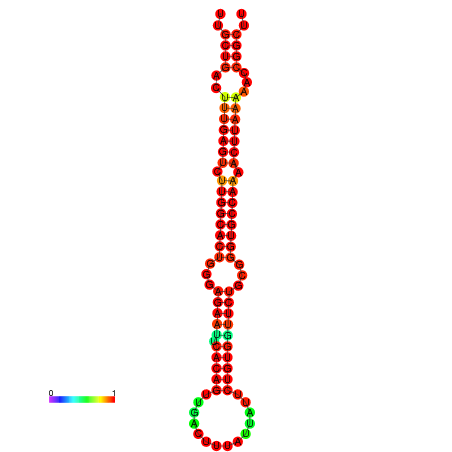
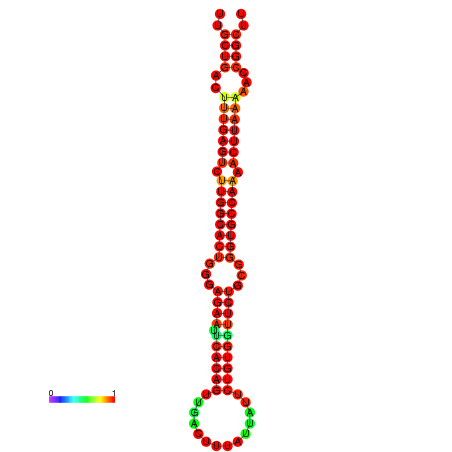
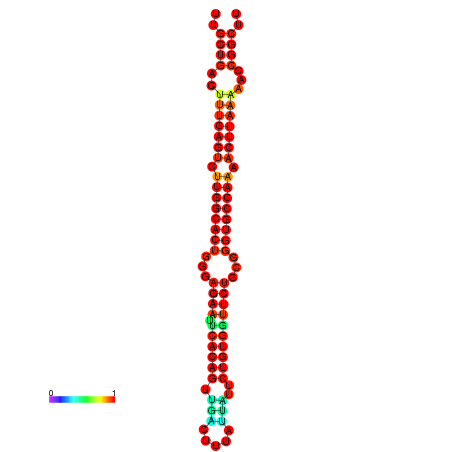
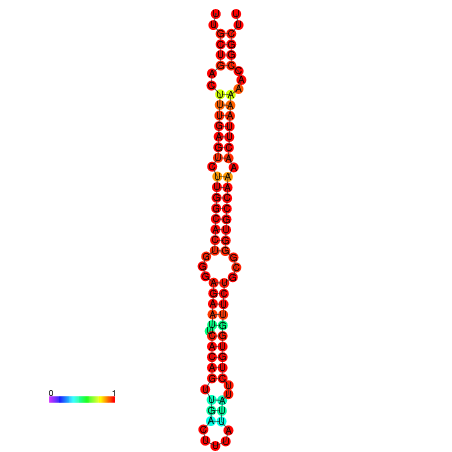
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:15836491-15836610 - | CTGTAGCTCT-----------------------TCAA----------------T------TTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTG-GTTCTGCGGGTGCC-AAAACTTAAAA------ACCG-GCTTTGTACAACGATTTCT------------ |
| droSim1 | chr3L:15181316-15181435 - | CTCTAGCTCT-----------------------TCAA----------------T------TTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTG-GTTCTGCGGGTGCC-AAAACTTAAAA------ACCG-GCTTTATACAACGATTTCT------------ |
| droSec1 | super_0:7944386-7944505 - | CGCTAGCTCT-----------------------TCAA----------------T------TTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTG-GTTCTGCGGGTGCC-AAAACTTAAAA------ACCG-GCTTTATACAACGATTTCT------------ |
| droYak2 | chr3L_random:2539138-2539257 - | CGATAGCTCT-----------------------TCAA----------------T------CTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTG-GTTCTGCGGGTGCC-AAAATTTAATC------AACG-GCTCTATACAACGATTTCT------------ |
| droEre2 | scaffold_4784:18090156-18090270 - | CTG--GCGAT-----------------------TCAA----------------ACCA---TAGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTG-GTTCTGCGGGTGCC-AAAATTTAATC------AACA-GCTCCATACAACG------------------ |
| droAna3 | scaffold_13337:11320710-11320858 + | TTGTGATTTT-----------------------TGAATCAAATATTCTGG-----ACTCTCTGCTGTCTTTGAATCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTG-GTTCTGCGGGTGCC-AAAATTGAAAT------AATG-GCCTAAAACCTCAAAAAAGGATATTTATTAT |
| dp4 | chrXR_group6:3598033-3598148 - | CGATCGTTA---------------------------------------------------TTGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTCCA---TATATTCTGTG-GTTCTGCGGGTGCC-AAAATCGAATG------TTCG-GCTCGAAACAGCAGCTGAA------------ |
| droPer1 | super_9:1894031-1894138 - | T------------------------------------------------------------TGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTTCA---TATATTCTGTG-GTTCTGCGGGTGCCCAAAATCGAATG------TTCG-GCTCGAAACAGCAGCTGAA------------ |
| droWil1 | scaffold_180727:1452378-1452537 + | CTTAAATTCTAAGAAAGATTTTCTGTGTGTGTTTGAA----------CGAAAA-TAAAGTCTGCTCACAATGCATCTTGGCACTGGGAGAATTCACAGTTGATTCTTATTCGATATTCTGTG-GTTCTGCGGGTGCC-AAAATCTAATG------TTTGAGCTGTGATCATCGAAAAAG------------ |
| droVir3 | scaffold_13049:24412507-24412633 - | TTGT-GTTCT-----------------------TCTA----------CGGAAA-GCAGAGCTACTGCCATTTCATCTTGGCACTGGGAGAATTCACAGTCGATTACAT---AATATTCTGTG-GTTCTGCGGGTGCC-AAAATGTAATG------TGTG-GACCAAGACAGC-----CT------------ |
| droMoj3 | scaffold_6473:2363995-2364108 + | ATGTAAATGT------------------------TAA----------------T------TAACTGAC------------CGTTGAGGGCATTTACATCTATGTATAA-----CAGTTTATGTAATATGCAAGTATC-TAAATATATACAGAAATACTA-TGTGTGTATAACAGTTTTT------------ |
| droGri2 | scaffold_15110:24319786-24319904 - | ATATTGTG-------------------------GAAA----------------G------CTACTGCCATTACATCTTGGCACTGGGAGAATTCACAGTTGGTTATAT---AATATTCTGTG-GTTCTGCGGGTGCC-AAACTGTAATG------TGTG-GACATAGA-AACAACCTGA------------ |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:15836491-15836610 - | CTGTAGCTCTTCAA-----------------------------------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTGTACAACGATTTCT------------ |
| droSim1 | chr3L:15181316-15181435 - | CTCTAGCTCTTCAA-----------------------------------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTATACAACGATTTCT------------ |
| droSec1 | super_0:7944386-7944505 - | CGCTAGCTCTTCAA-----------------------------------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTATACAACGATTTCT------------ |
| droYak2 | chr3L_random:2539138-2539257 - | CGATAGCTCTTCAA-----------------------------------TCTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTTAATCAACG-GCTCTATACAACGATTTCT------------ |
| droEre2 | scaffold_4784:18090156-18090270 - | CTG--GCGATTCAA-----------------------------ACC---ATAGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTTAATCAACA-GCTCCATACAACG------------------ |
| droAna3 | scaffold_13337:11320710-11320858 + | TTGTGATTTTTGAATCAAATATTCTG--------------G----ACTCTCTGCTGTCTTTGAATCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTGAAATAATG-GCCTAAAACCTCAAAAAAGGATATTTATTAT |
| dp4 | chrXR_group6:3598033-3598148 - | CGATCGTTA-----------------------------------------TTGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTCCA---TATATTCTGTGGTTCTGCGGGTGCC-AAAATCGAATGTTCG-GCTCGAAACAGCAGCTGAA------------ |
| droPer1 | super_9:1894031-1894138 - | T--------------------------------------------------TGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTTCA---TATATTCTGTGGTTCTGCGGGTGCCCAAAATCGAATGTTCG-GCTCGAAACAGCAGCTGAA------------ |
| droWil1 | scaffold_180727:1452378-1452537 + | CTTAAATTCTAAGA-AAGATTTTCTGTGTGTGTTTGAACGAAAATAAAGTCTGCTCACAATGCATCTTGGCACTGGGAGAATTCACAGTTGATTCTTATTCGATATTCTGTGGTTCTGCGGGTGCC-AAAATCTAATGTTTGAGCTGTGATCATCGAAAAAG------------ |
| droVir3 | scaffold_13049:24412507-24412633 - | TTGT-GTTCTTCTA------------------------CGGAAAGCAGAGCTACTGCCATTTCATCTTGGCACTGGGAGAATTCACAGTCGATTACAT---AATATTCTGTGGTTCTGCGGGTGCC-AAAATGTAATGTGTG-GACCAAGACAGC-----CT------------ |
| droGri2 | scaffold_15110:24319786-24319904 - | ATATTGT--GGAAA-----------------------------------GCTACTGCCATTACATCTTGGCACTGGGAGAATTCACAGTTGGTTATAT---AATATTCTGTGGTTCTGCGGGTGCC-AAACTGTAATGTGTG-GACATAGA-AACAACCTGA------------ |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| -33.2 | -33.2 | -33.0 | -33.0 | -32.2 | -32.2 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -33.2 | -33.2 | -33.0 | -33.0 | -32.2 | -32.2 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -33.2 | -33.2 | -33.0 | -33.0 | -32.2 | -32.2 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
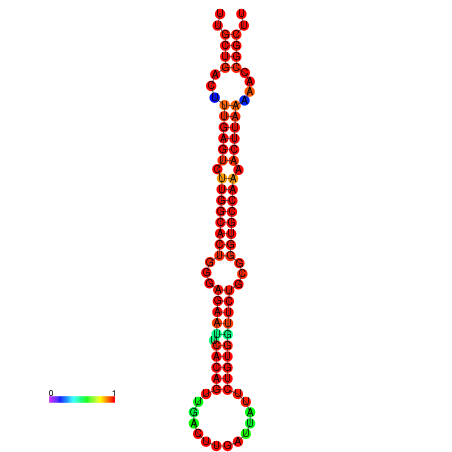 |
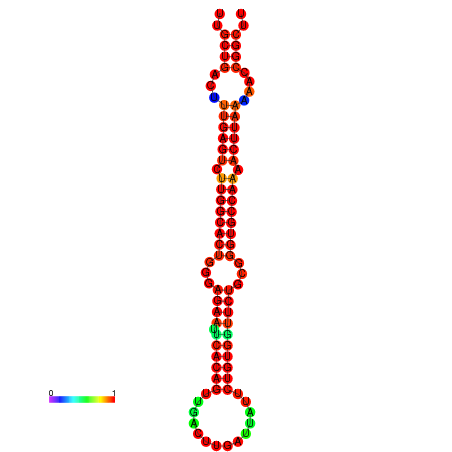
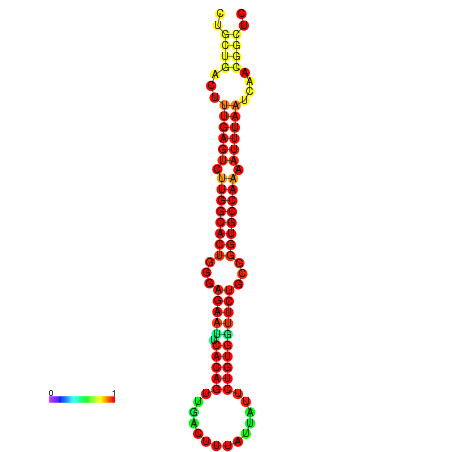
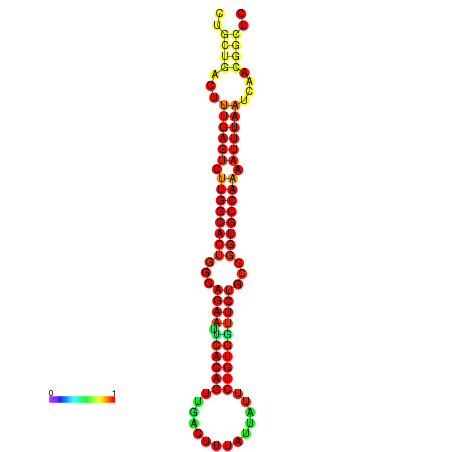
| -30.5 | -30.5 | -30.3 | -30.3 | -29.5 | -29.5 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -33.5 | -33.5 | -32.9 | -32.9 | -32.8 | -32.8 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -29.5 | -29.5 | -28.9 | -28.9 | -28.8 | -28.8 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -32.3 | -32.3 | -32.3 | -32.3 | -31.3 | -31.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -29.0 | -29.0 | -29.0 | -29.0 | -28.8 | -28.8 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -29.9 | -29.9 | -29.8 | -29.8 | -29.6 | -29.6 |
|---|---|---|---|---|---|
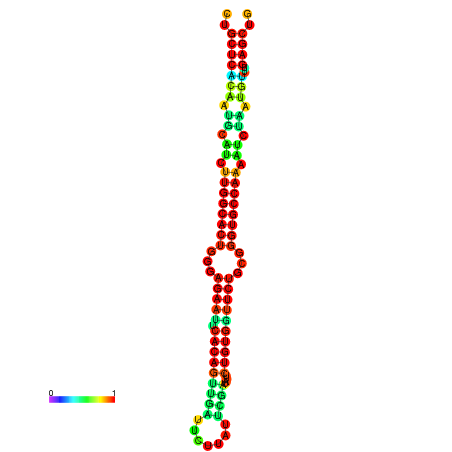 |
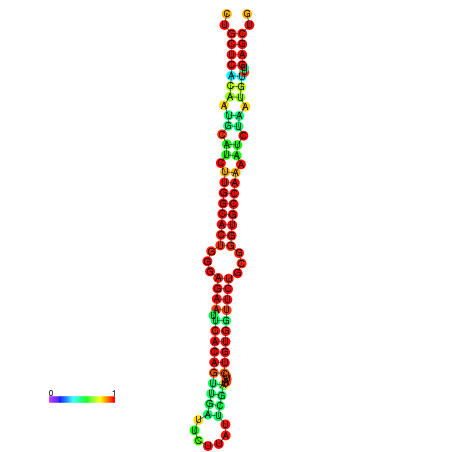 |
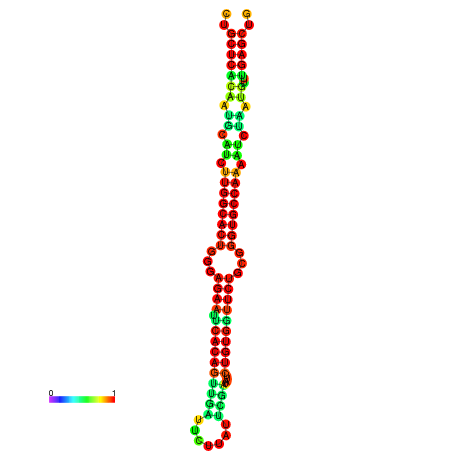 |
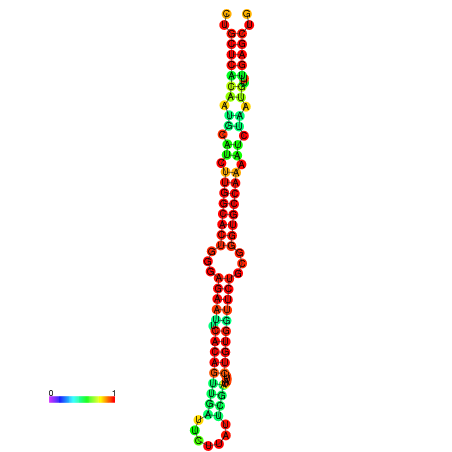 |
 |
 |
| -32.6 | -32.6 | -32.2 | -32.2 | -32.0 | -32.0 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -10.1 | -10.0 | -9.9 | -9.8 | -9.7 | -9.7 |
|---|---|---|---|---|---|
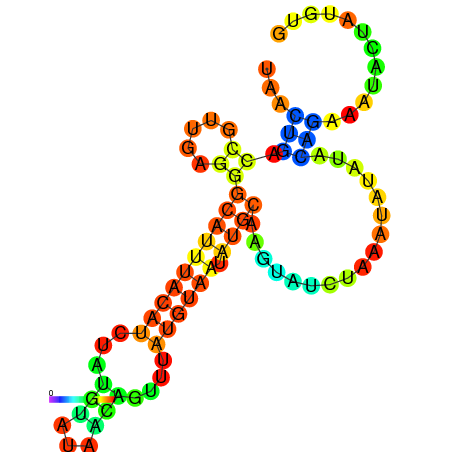 |
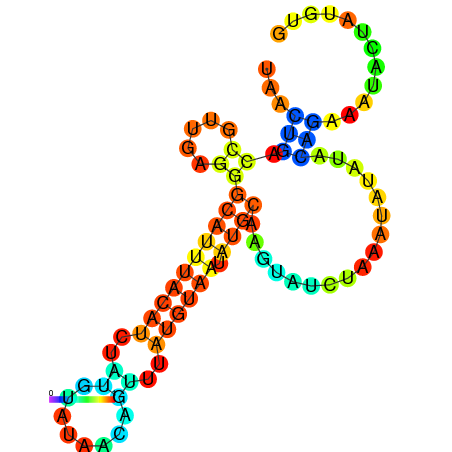 |
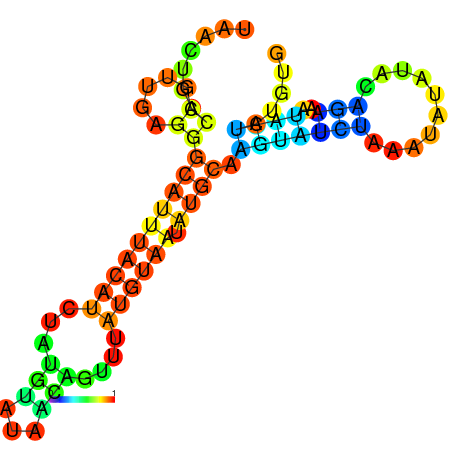 |
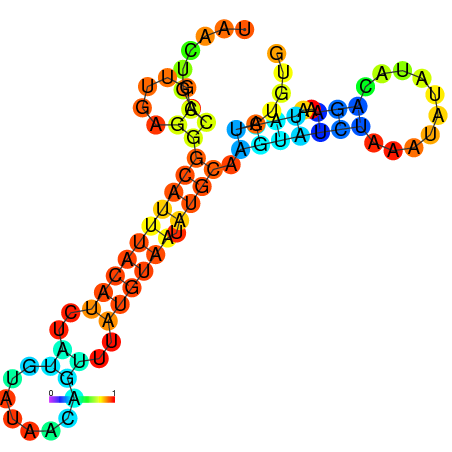 |
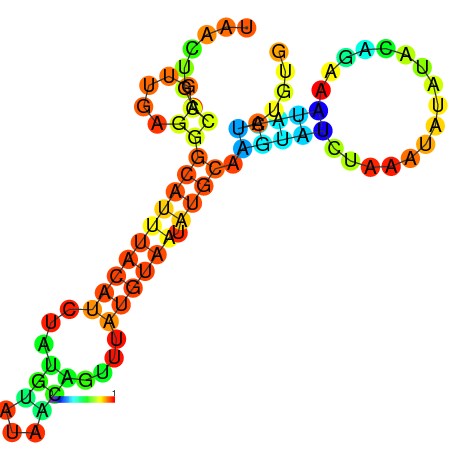 |
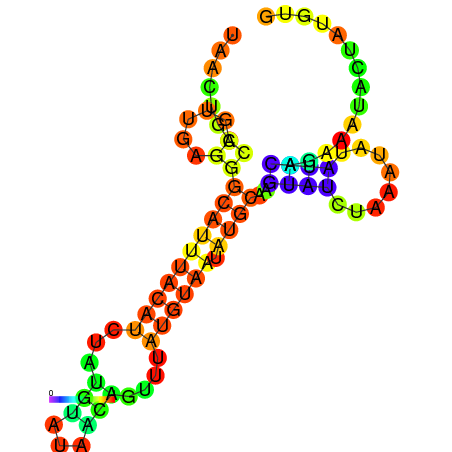 |
| -36.0 | -36.0 | -35.4 | -35.4 | -35.2 | -35.2 |
|---|---|---|---|---|---|
 |
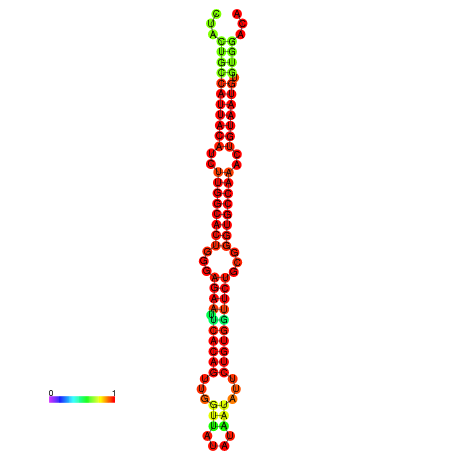 |
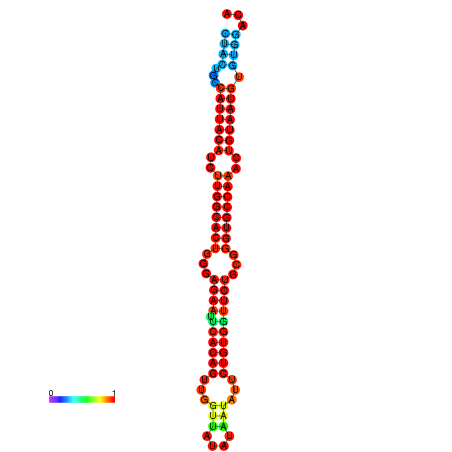 |
 |
 |
 |
Generated: 01/05/2013 at 05:12 PM