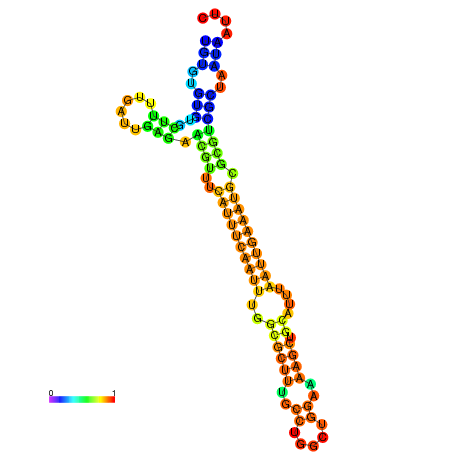
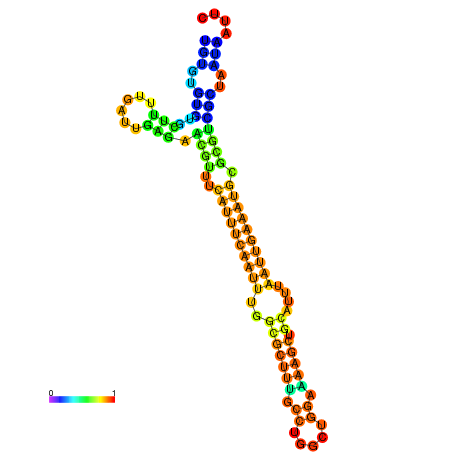
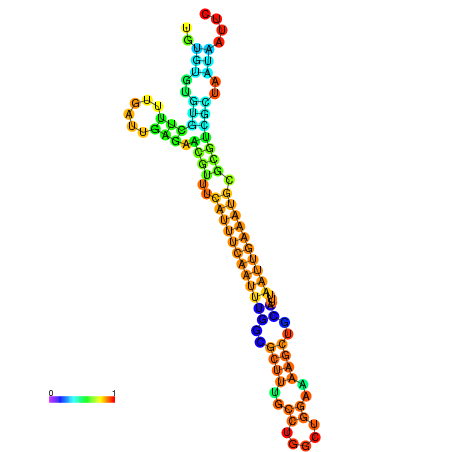
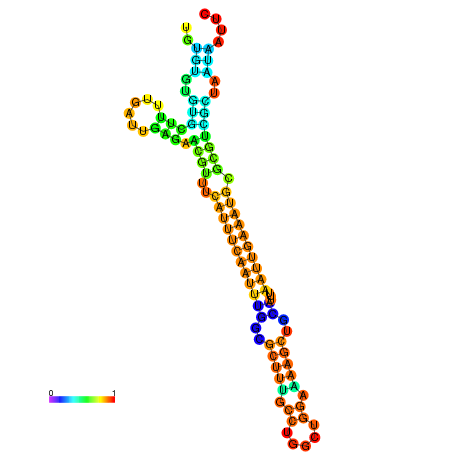
| dm3 |
chr2L:5068555-5068699 + |
CGTATGTAT---------------------TATACATGTATGATCACATTGGCTTGTGGGCGTGG--CAC-TTCAATTGGGCTGACTTTGCCTGGCTGGAAAAGCGGCA-CCTAATTGAAATGC-CCGC---CGATCAGT-GCTCGATTACGAT------CGACACAAGCGACATT---TTCGT----------------- |
| droSim1 |
zds89d09.b1:77-211 + |
CATACGTACAT----------------------ACATGTATGATC-CATTGGCTTGTGGGCGCGG--CAC-TTCAATTGGGCTGACTTTGCCTGGCTGGAAAAGCGGCA-CCTAATTGAAATGC-CCGC---CAATC---------------------------------GACATT---TTCGTTTAATTTCGTTTCTAGC |
| droSec1 |
super_5:3155468-3155580 + |
CGTACAT--------------------------ACATGTATGATC-CATTGGCTTGTGGGCGTGG--CAC-TTCGATTGGGCTGACTTTGCCTGGCTGGAAAAGCGGCA-CCTAATTGAAATGC-CCGC---CAATC---------------------------------GACATT---TTCGT----------------- |
| droYak2 |
chr2L:10456243-10456376 - |
TATAC---------------------------------TATAAT--CATTGGCTTGTGGGCGTGG--CAC-TTCAATTGGTCTGACTTTGCCCGACTGGAAAAGCGGCA-CCTAATTGAAATGC-CCGCCA-CAATCAGT-GCTCGATCGTGAT------CGACACAAGCGACATT---TTCGT----------------- |
| droEre2 |
scaffold_4929:5148425-5148554 + |
TTTATG------------------------------------ATC-CATTGGCTTGTGGGCGTGG--CAC-TTCAATTGGTCTGACTTTGCCCGGCTGGAAAAGCGGCA-CCTAATTGAAATGC-CCGC---CAAACAGT-GCTCGACCATGAT------CGGCACAAGCGACATT---TTCGT----------------- |
| droAna3 |
scaffold_12916:13706207-13706309 + |
CATATG-----------------------------------------------TTGTGGGCGTGG--CAC-TTCAATTGGTCTAACTTTCCCGGAATGGTAAAGCAGTCACTTAATTGAAATGC-CCGC---CAATC---------------------------ACGAGCAACATTTTTTTCGT----------------- |
| dp4 |
chr4_group4:1588263-1588389 + |
TGTCTGTGTGCG----------------------------------CATTGGGTTG-GGGCGTGG--CAC-TTCAATTTGGC--GCTTTGCCTGGCTGTAAAAGCTGCA-ACTAATTGAAATGT-CCGC---CAATCAATTAAGCAA---TAAA------CAACATAAGCAACACT---TTCGA----------------- |
| droPer1 |
super_10:591769-591895 + |
TGTCTGTGTGCG----------------------------------CATTGGGTTG-GGGCGTGG--CAC-TTCAATTTGGC--GCTTTGCCTGGCTGTAAAAGCTGCA-ACTAATTGAAATGT-CCGC---CAATCAATTAAGCAA---TAAA------CAACATAAGCAACAAT---TTCAA----------------- |
| droWil1 |
scaffold_180703:1359544-1359639 + |
TTCACAC----------------------------------------ATTCGTTTTTGTCTATGG--TATTTTCAATTTGGCTC-CTTTATCTGGCTGAAAAAGCTGCA-ACTAATTGAAATGC-CCAT---TTAAC-------------------------------------TT---TATAT----------------- |
| droVir3 |
scaffold_12963:10059299-10059444 - |
CGTATGTGTGTGTGTGTGTGTGTCTGTGTCTGTGTTTGTGTGTGTGCTTTTGATTGAGAACGTTT--CAT-TTCAATTTGGC--GCTTTGCCTGGCTGGAAAAGCTGCA-TTTAATTGAAATGC-GCGTCGCTAAT-AA---TTC-------------------------AATAATT--TATTA----------------- |
| droMoj3 |
scaffold_6500:1220428-1220559 + |
TTTGAGT----------------------------------------ATCA-TTTTAGGGCGTGTTCCAT-TTCAATTTGGC--GCTTTGCCTGGCTGGAAAAGCTGCA-TTTAATTGAGATGC-GCGT---CGACATTTTATTCGATTACATTTAGTTTAATTACAAGAAATATT---TAAGC----------------- |
| droGri2 |
scaffold_15252:11801593-11801729 + |
TGTGTGTGTGTG--------CTTCTCTCTCTATCTCTGTATCTCTCTCTCTGGTTGAGGGCGTTC--CAT-TTCAATTTGGCGCTTTTTGTCTGGCTGGAAAAGCTGCACATTAATTGAAATGTCGCGTCGTTAATCAACT--------------------------------AAT---TT-TG----------------- |