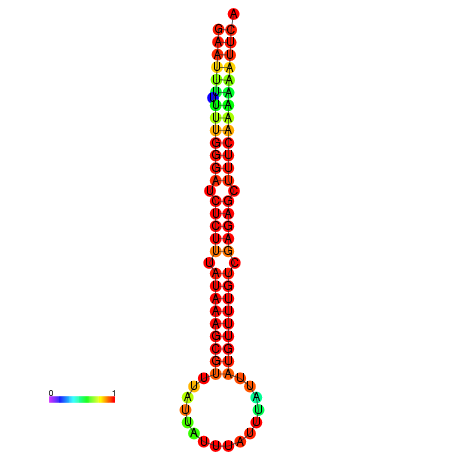
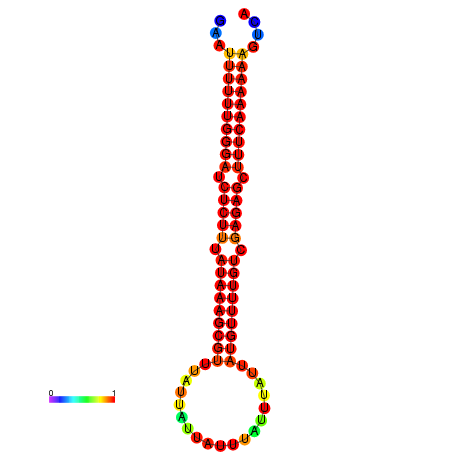
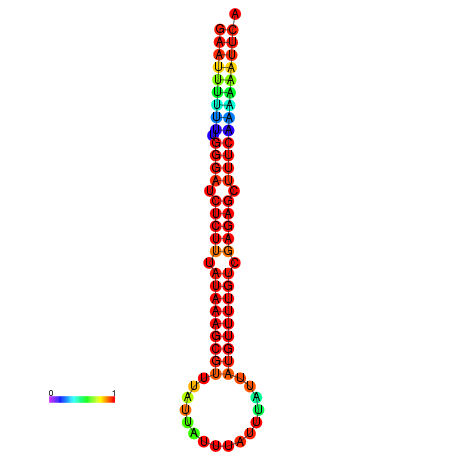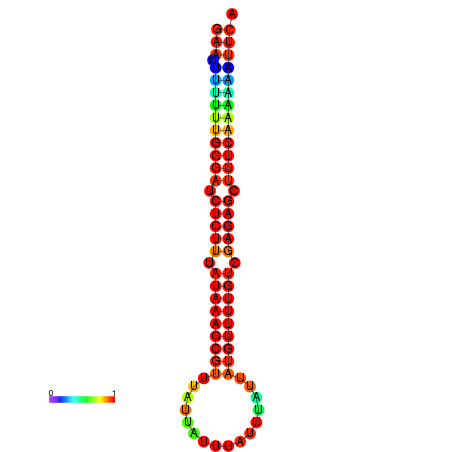| dm3 |
chrX:11441440-11441545 + |
ACAAAATTCACACTCGAAT-TTTTTTGGGATCTCTTTATAAAGCGTTTATTA---TTTATTTATTATGTTTTGTCGAGAGCTTTCAAAAA-ATT-CAGAAGACAACAACGCA |
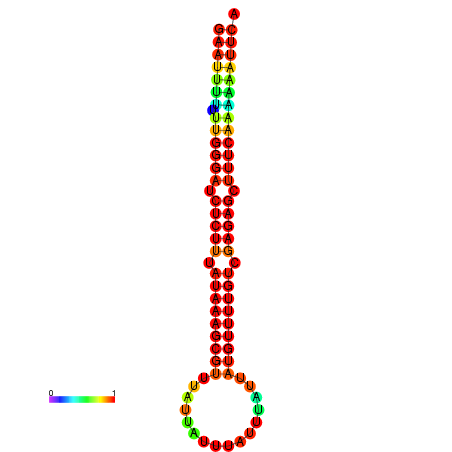
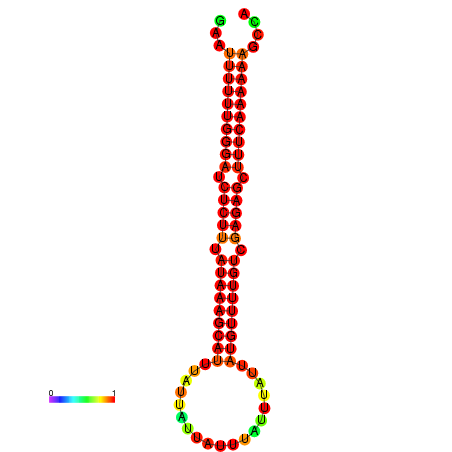
| droSim1 |
chrX_random:3203326-3203430 + |
ACAAAATTCACACTCGAAT-TTTT-TGGGATCTCTTTATAAAGCGTTTATTATTATTTATTTATTATGTTTTGTCGAGAGCTTTCAAAAA-AGT-CAGAAG---ACAACGCA |
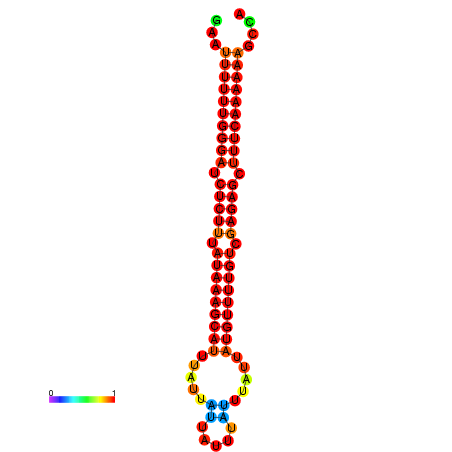
| droSec1 |
super_36:155641-155745 + |
ATAAAATTCACTCTCGAAT-TTTT-TGGGATCTCTTTATAAAGCATTTATTATTATTTATTTATTATGTTTTGTCGAGAGCTTTCAAAAA-AGC-CAGAAG---ACAACGCA |
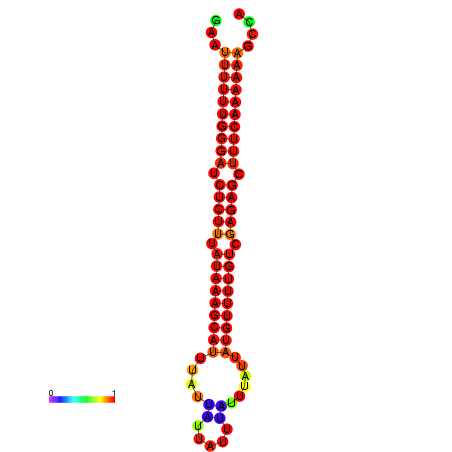
| droYak2 |
chrX:10720999-10721105 - |
ACAAAATTCACACTCGAAT-TTTTTGGCAATCTGTTTATAAAGCGTTTATTA---TTTATTTATTATGTTTTGTCGAGAGCTTTCAAAAATACA-ACAAAATAGGAAAAAAA |
| droEre2 |
scaffold_4690:3720646-3720750 - |
ACAAAATTCACACTCGAATTTT---GGGTATCTCTTTATAAAGCGTTTATTA---TTTATTTATTATGTTTTGTCGAGAGCTTAAAAAAA-ATATGAAAAGACAACAACCAA |
| droAna3 |
scaffold_13047:188962-189053 + |
ACAAAATTCACACTCGCTT-TT---GGCCAAC--TCCATAAAGCGTTAATTA---TCTG---------GCATTCTGAGAGCCAAAAGCTA-AAT-CAAATTCGATCGGAGCG |
| dp4 |
chrXL_group1e:1762210-1762257 + |
ACAAAATTCACACTCGAAT-TT---GGGGATCGCGTAGCGAGG--------------------------------------------------------GG----GGGGCAA |
| droPer1 |
super_18:601670-601716 + |
ACAAAATTCACACTCGAAT-TT---GGGGATCGCGTAGCAAGGG-------------------------------------------------------------GGGGCAA |
| droWil1 |
scaffold_180777:122952-122984 + |
ATAAACTCAATACACGAAT-TTGGTACTGATTTC------------------------------------------------------------------------------ |
| droVir3 |
Unknown |
---------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6500:22511787-22511827 + |
TCC---------------------------------------------------------------------GTCAATCATCTCCTTAAA-ACT-CTAAAGCAAACCAAGCA |
| droGri2 |
scaffold_15203:463820-463836 - |
GTGAAATTCACACTCGA----------------------------------------------------------------------------------------------- |