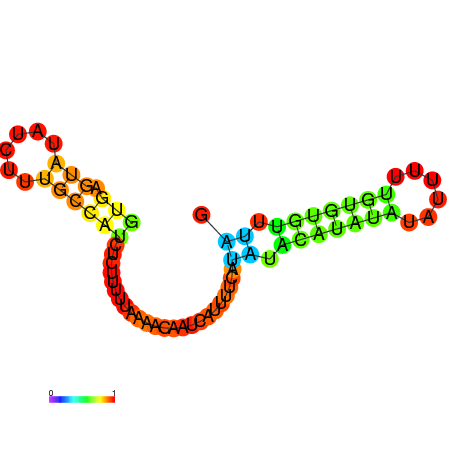
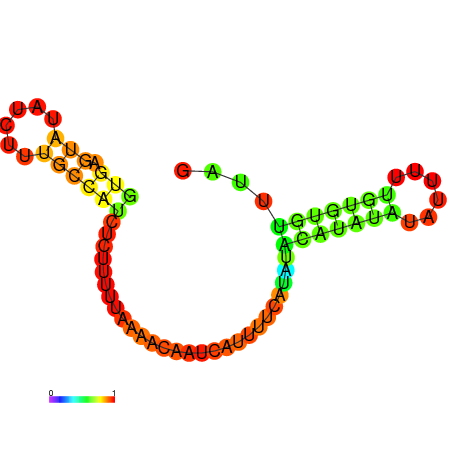
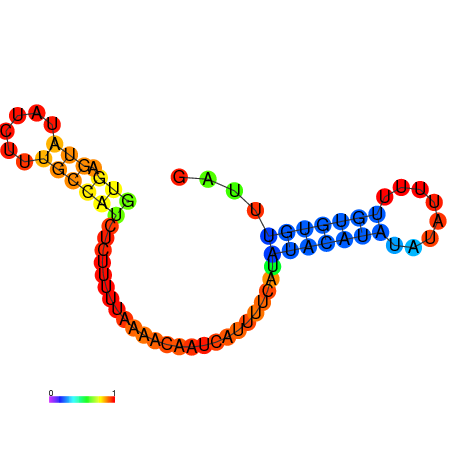
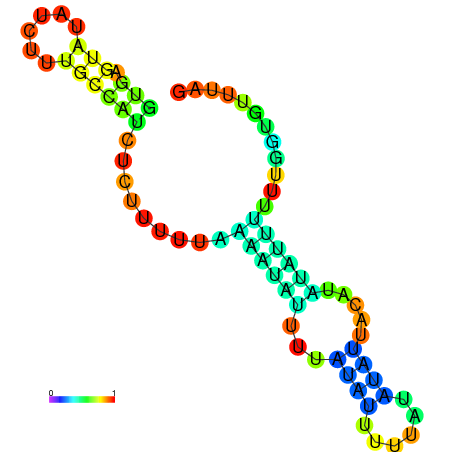
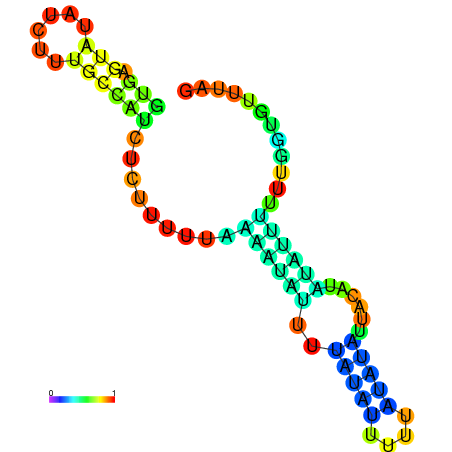
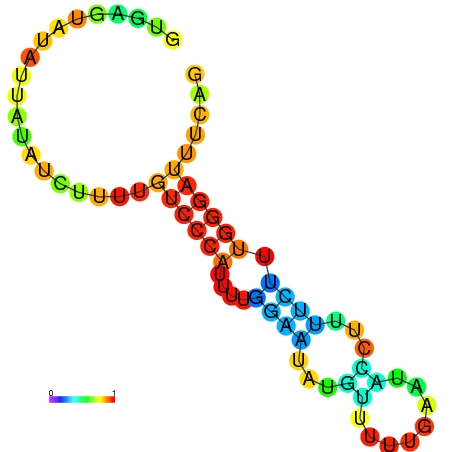
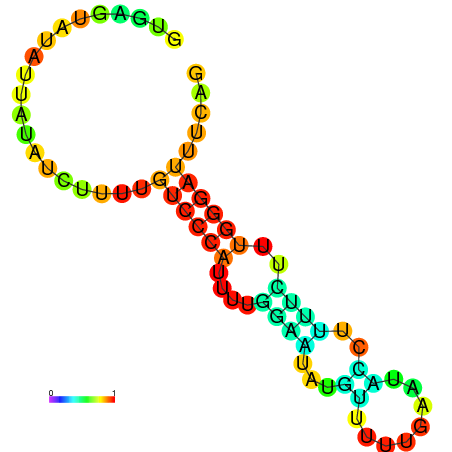
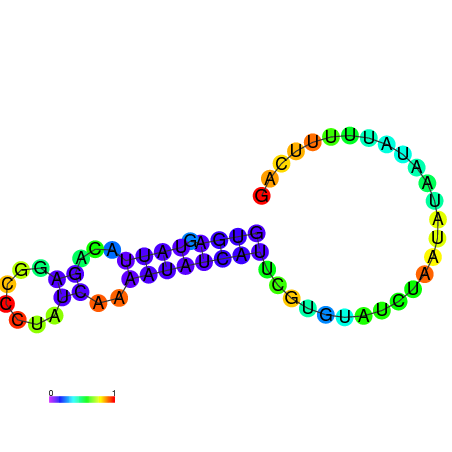
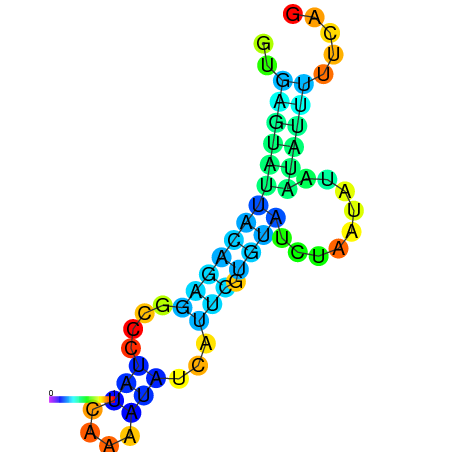
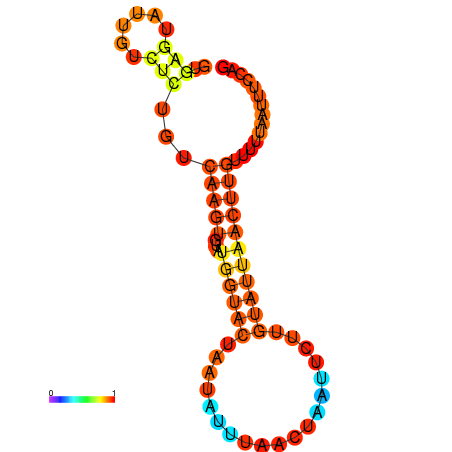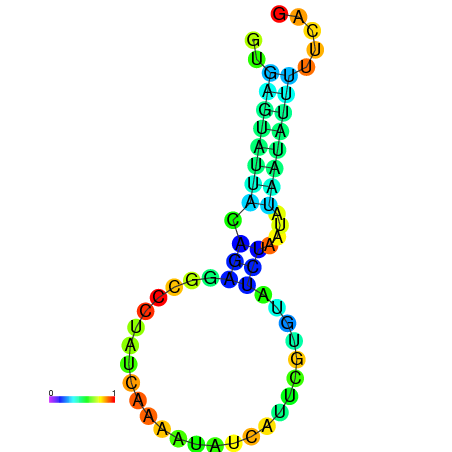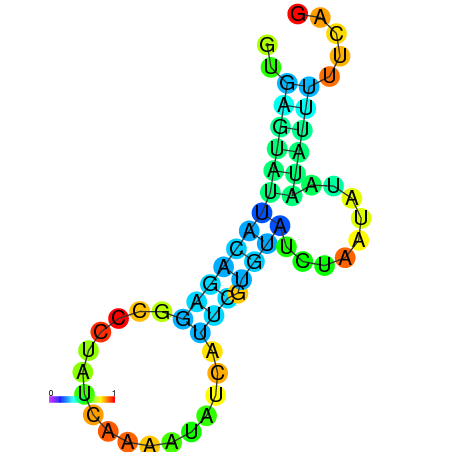| dm3 |
chr2L:13609973-13610070 + |
AGCAATAAGTTAGACGTGAGTATG------GCTATACAAAT---------CATCT-------GGAACGT-------AAGTTCTAATG-----TATGTATTG---TATG-----TTGTATTTGCAGTTTTATGATAGCAAA |
| droSim1 |
chr2L:13379253-13379350 + |
AGCAATAAGTTAGACGTGAGTATG------GCTATACAAAT---------CATCT-------GGAACAT-------ACGTTCTAATG-----CATGTATTG---TATG-----TTGTATTTGCAGTTTTATGATAGCAAA |
| droSec1 |
super_16:1746050-1746147 + |
AGCAATAAGTTAGACGTGAGTATG------GCTATACAAAT---------CATCT-------GGAACAT-------ACGTTCTAATG-----CATGTATTG---TATG-----TTGTATTTGCAGTTTTATGATAGCAAA |
| droYak2 |
chr2L:10029706-10029801 + |
AGCAATAAGTTAGATGTGAGTATG------GCTATTTAAAT---------CAACC-GAAC--GT-ATGT-------ATGTTATATAG-----T-----TTG---CATT-----TTGTATTTGCAGTTTTATGATAGCAAA |
| droEre2 |
scaffold_4929:11847836-11847927 - |
AGCAATAAGTTAGACGTGAGTATG------GCTATCTAAAT---------CAGCC--------GAACGT-------GTATTCTAATG-----TTTG--------CACT-----TTGTATTTGCAGTTTTATGATAGCAAA |
| droAna3 |
scaffold_12943:1669999-1670103 + |
AGCGATAAGTTAGACGTGAGTATT-----GTCTCTGTCAAG---------TGTAT-------GGTACTAATATTTA----ACTAATT-----CTTGTATTAACTTGTT-----TTTAATTTCCAGTTTTATGATAGCAAT |
| dp4 |
chr4_group1:1879331-1879430 - |
AGCGATAAGTTAGACGTGAGTATA------TCTTTGCCATC---------TCTTT-----TTAAAACAA-------TCATTTTCATA-----TACATATAT---ATTT-----TTGTGTGTTTAGTTTTATGATAGCAAC |
| droPer1 |
super_5:3326174-3326277 + |
AGCGATAAGTTAGACGTGAGTATA------TCTTTGCCATC---------TCTTT-----TTAAAATAT----TTTATATTTTTATATATTACATATATTT------------TTTGGTGTTTAGTTTTATGATAGCAAC |
| droWil1 |
scaffold_180772:852551-852648 - |
AGCGATAAGTTAGATGTGAGTATA------T-TATATCTTTTGTCCCATT----------TTGGAATAT-------GTTTTTGAATA-----CCTT--------TTCT-----TTGGGATTTCAGTTTTATGATAGCAAC |
| droVir3 |
scaffold_12723:5236355-5236448 - |
AGTGATAAGTTAGACGTGAGTATT------ATCAACATATATGAATTATT----------T-TAAAATA-------ATAATC-------------GAAT-T---TAAT-----TTGTTATCCCAGTTTTATGATAGCAAC |
| droMoj3 |
scaffold_6500:16444108-16444197 + |
AGTGATAAGTTAGACGTGAGTATTACAGAGGCCCTATCAA------------------------AAT-A-------TC------ATT-----CGTGTATCT---AATATAATAT----TTTTCAGTTTTATGATAACAAC |
| droGri2 |
scaffold_15126:8263493-8263587 - |
AGTGATAAGTTAGACGTGAGTATTT-----ATTAAC--------------TTCATTCAAC--GTAATAT-------ACATACTAAAA-----TATGAA--------TACATCAT----TTTGCAGTCTTATGATAGCAAC |