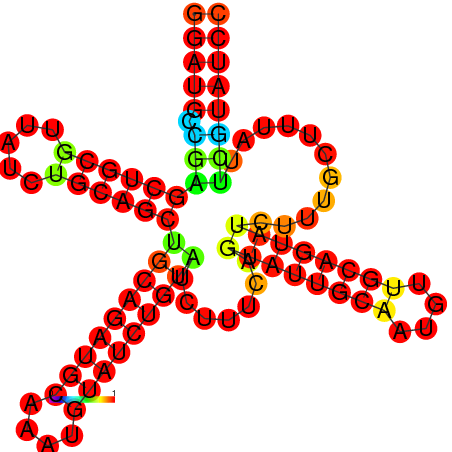
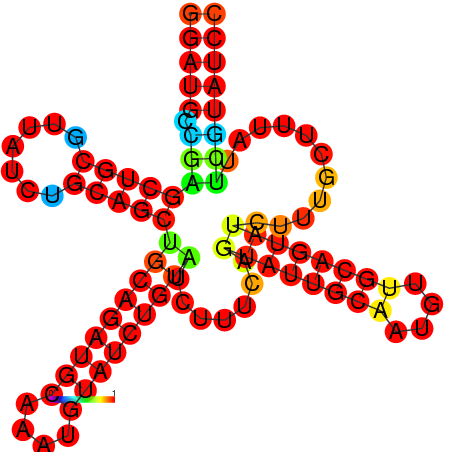
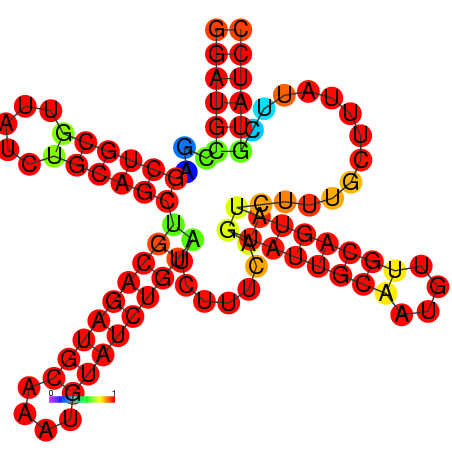
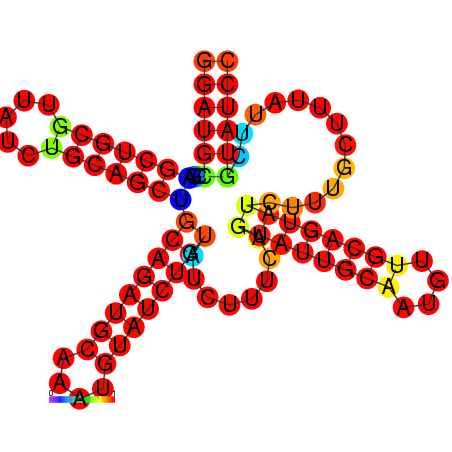
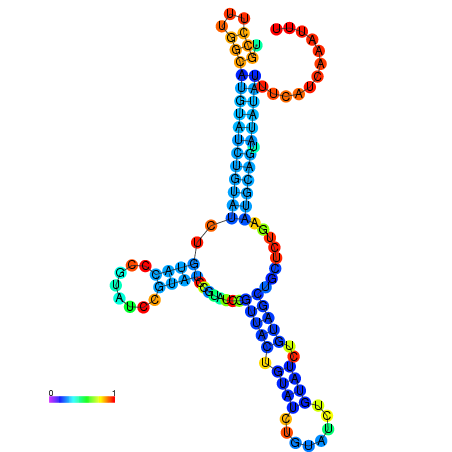
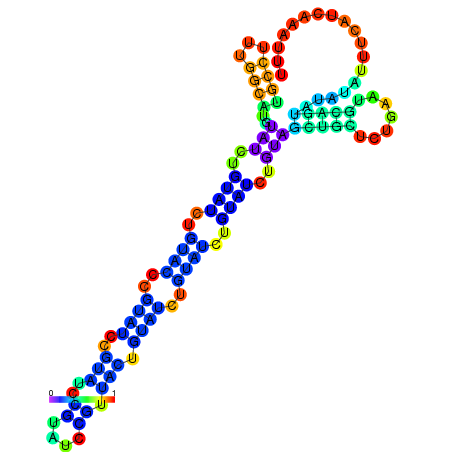
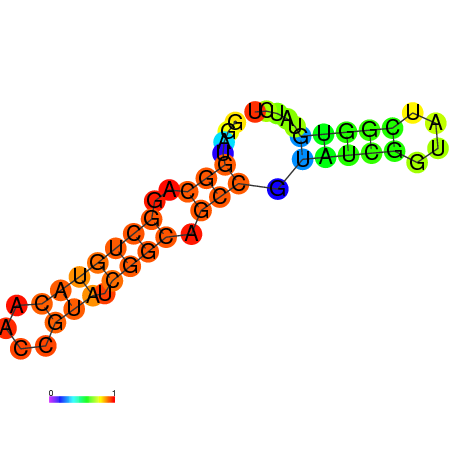
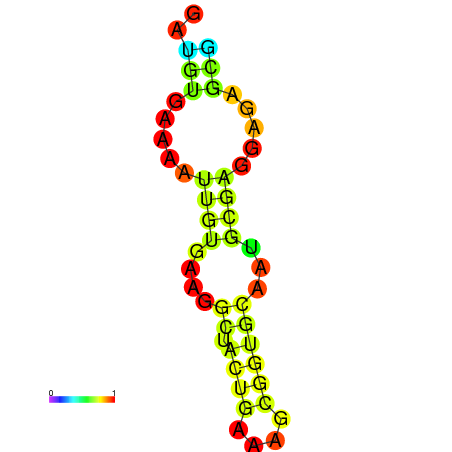
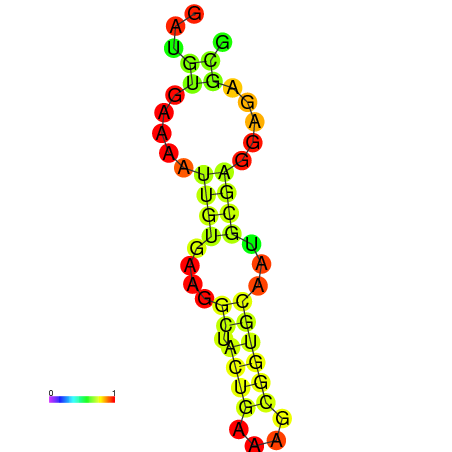
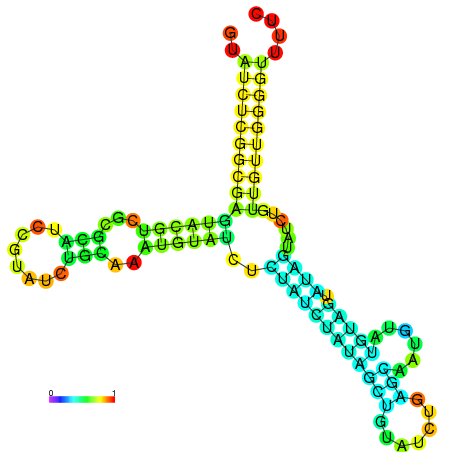
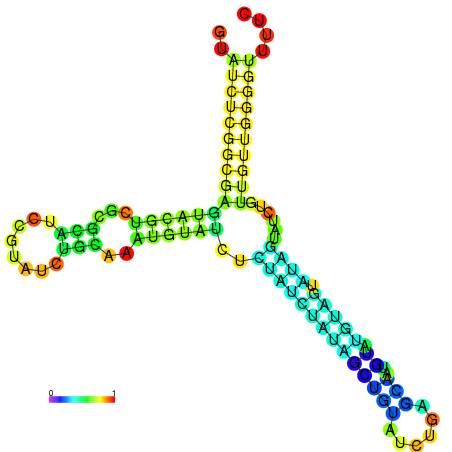
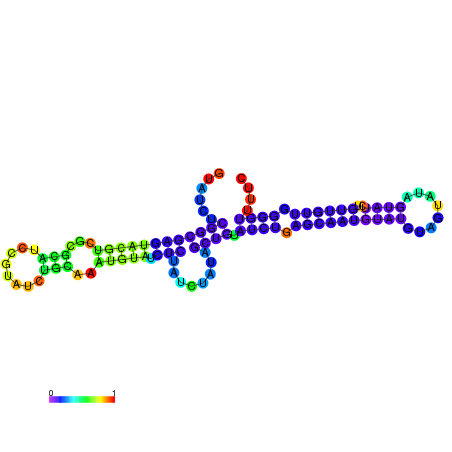
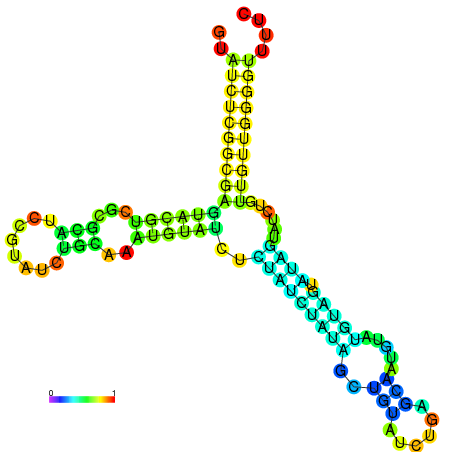
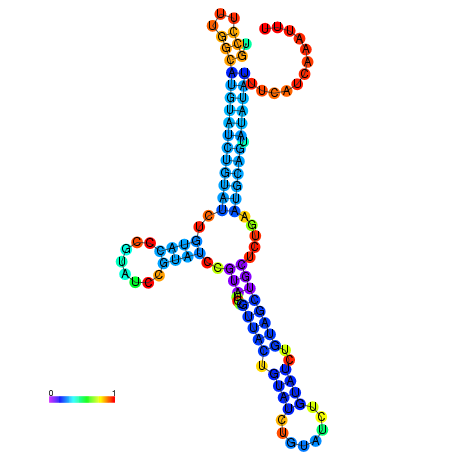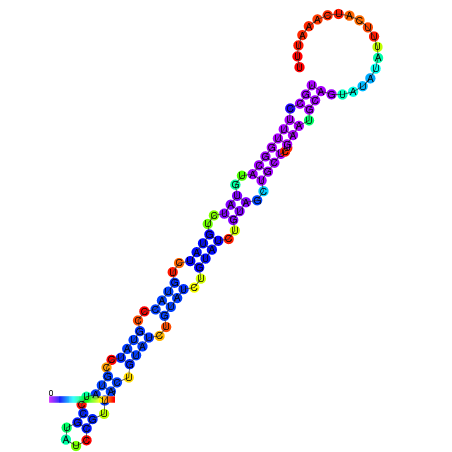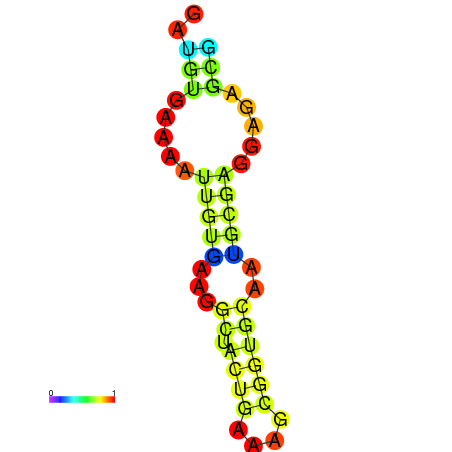| dm3 |
chr3R:10222106-10222238 - |
TCCAGTG-------------------------------------------------------------------------CATCCTACGGATGTCGAGCAGTACACGTA---TTATCTGCAG------CTGCAGATGCAAATGTATATGTATCTGTATCTG------TATCT--GCAGTAT-TGC-----AGTA-------TCGCTGCTTTATTCATATCCATG-------AGACGCGTCGCG |
| droSim1 |
chr3R:11233896-11234028 + |
GCCAGTG-------------------------------------------------------------------------CATCCTACGGATGTCGAGCAGTACTCGTA---TTATCTGCAG------CTGCAGATGCAAATGTATATGTATCTGTATCTG------TATCT--GCAGTAT-TGC-----AGTA-------TCGCTGCTTTATTCGTATCCATG-------TGACGCGTCGCG |
| droSec1 |
super_0:11818438-11818570 + |
GCCAGTG-------------------------------------------------------------------------CATCCTACGGATGTCGAGCAGTACTCGTA---TTATCTGCAG------CTGCAGATGCAAATGTATATGTATCTGTATCTG------TATCT--GCAGTAT-TGC-----AGTA-------TCGTTGCTTTATTCGTATCCATG-------TGACGCGTCGCG |
| droYak2 |
chr3R:16218838-16218961 + |
TCCAGTG-------------------------------------------------------------------------CATCCTACGGATGCCGAGCTGC------G---TTATCTGCAG------CTGCAGATGCAAATGTATCTGTATCTT--TCAG------TAT-T--GCAATGT-TGC-----AGTA-------TCTTTGCTTTATTCGTATCCGTG-------TGACGCGTCGCG |
| droEre2 |
scaffold_4770:9756449-9756546 - |
TCCAGTG-------------------------------------------------------------------------CATCCTACGGATGCCGAGCAT----C--G---TTATCTGCAG------CTGCAGATGCAAATGTA------------TCTG------TATCT--GCAGTAC-GGC-----------------------------TGTATCCGTG-------TGACGCGGCGCG |
| droAna3 |
scaffold_13117:2846333-2846460 - |
GTA----------------------------------------------------------------------------------------TCTCGAGATGCATCTGTAAGAGTATCTGCAT------CTGCAAATGTATCTGTATCTGTATCTGTATCTG------TATCT--GTATCTG-TGC-----AAGACTTGCCACCTCTCCTTTTCTCCTTTTCCTG-------AGCAAATGTCGC |
| dp4 |
chrXR_group8:8158458-8158593 + |
GCTAGTG-------------------------------------------------------------------------CCCACTACTGCCTTTGGCATGTATCTGTATCTGTACCCGTATCCGTATCCGTATCCGTTACTGTATCTGTATCTGTATCTG------TAGCT--GCTCTGAATGC-----AGTA-------TATA---TTTCATCAAATTTTTG-------TA----CTCGCA |
| droPer1 |
super_0:4124990-4125137 + |
GTAGGGGATCAGGCATGGCCAGGGGCAGCGGCAACGGCAGGGGCAGGGGCAGGGGCAGCTTTTCCAGGCT-------GTGCATCCTACGGATGGCAGGCTGTAC----------------------------------AACCGTATCGGCAGCCGTATCGG------TATCG--G---------------------------------------TGTATCTGTG-------TGACGCGTCGCG |
| droWil1 |
scaffold_181089:10073219-10073346 + |
CCTGGTAGAGAACGT-----------------------TGGGG--GAGGGGGGGGTTAGTTTTCCAGGCTCGCTGCAAAACATCCTACGATGTGAAAATTGT------G---A---------------------------AGG------------CTACTGAAA--GCG-GT--GCAATGC-GAG-----------------------------GAGAGCGACG-------CAATGCGACGCG |
| droVir3 |
scaffold_12928:2113057-2113188 + |
GTATCTATATCTGTA----------------------------------------------------------------------TATG------TATCTGT------ATCTGTATCTGTATCTGTATCTGTATCTGTATCTGTATCTGTATCTGTATCTG------TATCT--GTAT------C-----TATAT------TTGCTT----CTTTAAATCCGCCTCTTTCGAGACATTTATCC |
| droMoj3 |
scaffold_6654:2537235-2537365 + |
TTCAGTGAATCTGTATCTGTATCTGTATCTGTATCTG----------------------------------------------------------------------------TATCTGTAT------CTGTATCTGTATCTGTATCTGTATCTGTATCTGTATCTGTATCG--TATCTGT-ATC-----------------TGTATCTGTATCTGTATCTG----------TATTTGTATCT |
| droGri2 |
scaffold_14830:2155779-2155908 - |
GCGA-TG-------------------------------------------------------------------------TATCT--CGTATCTCGGCGAGTACGTCGC---------GCAT------CCGTATCTGCAAATGTATCTCTATCTATAGCTG------TATCTGAGCAATGTATGTAGTATAGTA-------TCTGTTGTT--GGGGTTTTCATA-------TTGCTTGTGCTG |