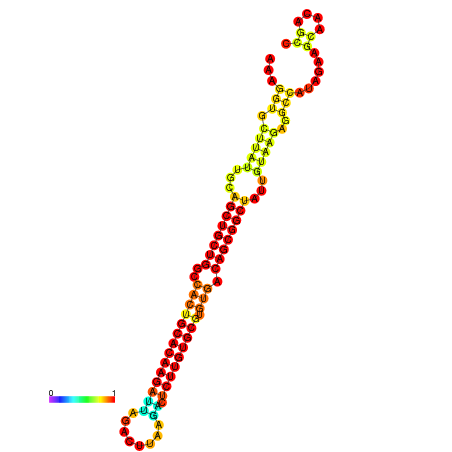
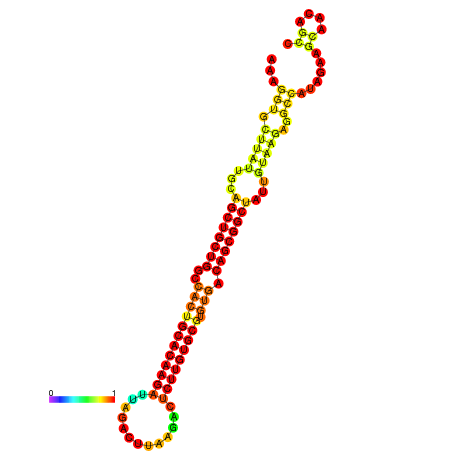
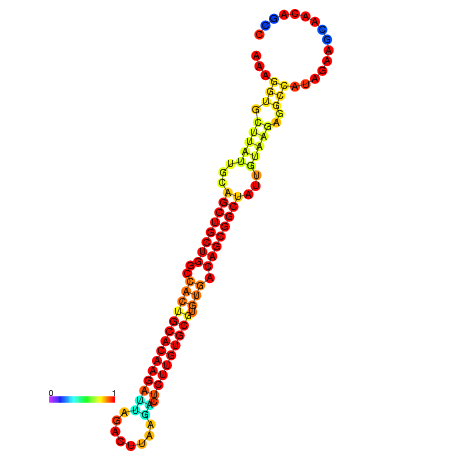
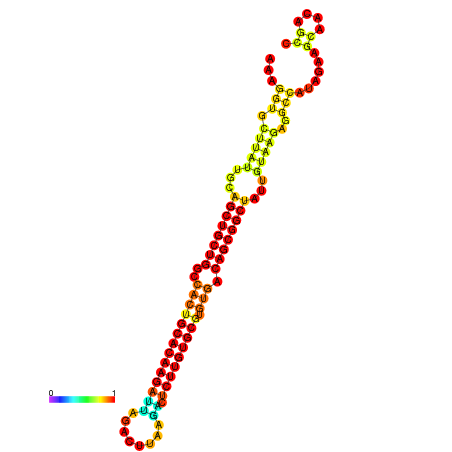
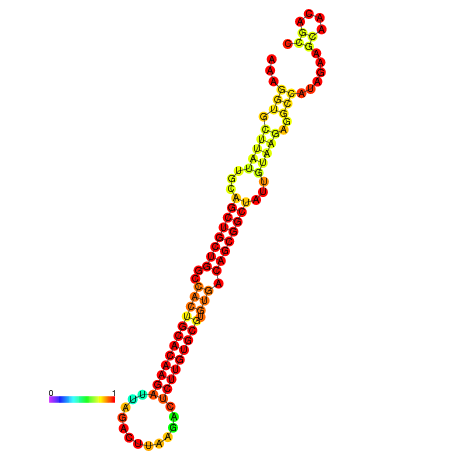
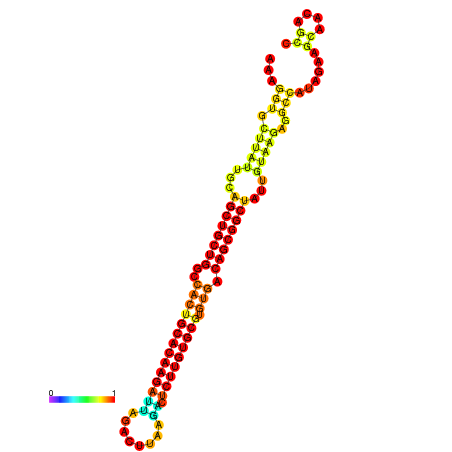
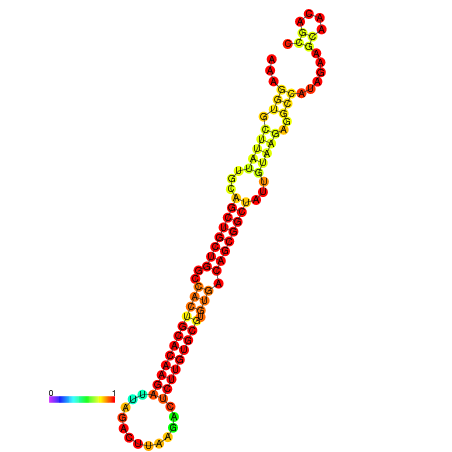
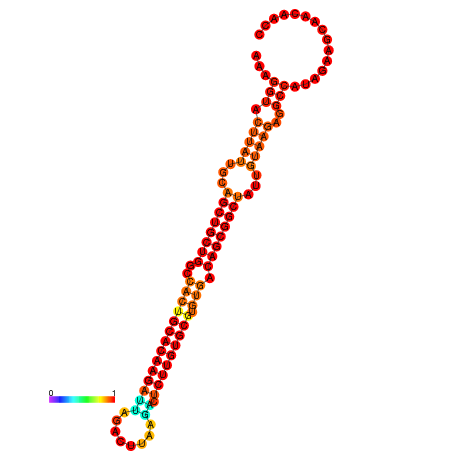
| ATATGT---GATTTAGCTCGATCGGTACTTATTGCAGCTGCTGGCCACTGCACAAGATTAGATTTAAGACTCTTGTGCGTGTGACAGCGGCTATTGTAAGAGGCCATATTAGCATCAAAACTAAACAAACCAAAAAGGAAGTTGAACGCCTAATGCAATCGATGAGTGTTGGGTTGCTTCAGTTTC | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| .......................................................................CTTGTGCGTGTGACAGCGGCTAT............................................................................................ | 23 | 1 | 53563.00 | 53563 | 23 | 53540 |
| .......................................................................CTTGTGCGTGTGACAGCGGCT.............................................................................................. | 21 | 1 | 47134.00 | 47134 | 8 | 47126 |
| ........................................................................TTGTGCGTGTGACAGCGGCTAT............................................................................................ | 22 | 1 | 42465.00 | 42465 | 25 | 42440 |
| ........................................................................TTGTGCGTGTGACAGCGGCT.............................................................................................. | 20 | 1 | 22563.00 | 22563 | 8 | 22555 |
| .......................................................................CTTGTGCGTGTGACAGCGGCTA............................................................................................. | 22 | 1 | 18745.00 | 18745 | 6 | 18739 |
| ........................................................................TTGTGCGTGTGACAGCGGCTA............................................................................................. | 21 | 1 | 18036.00 | 18036 | 7 | 18029 |
| ........................................................................TTGTGCGTGTGACAGCGGC............................................................................................... | 19 | 1 | 15366.00 | 15366 | 0 | 15366 |
| .......................................................................CTTGTGCGTGTGACAGCGGC............................................................................................... | 20 | 1 | 4416.00 | 4416 | 5 | 4411 |
| .......................................................................CTTGTGCGTGTGACAGCGG................................................................................................ | 19 | 1 | 2266.00 | 2266 | 0 | 2266 |
| .......................................................................CTTGTGCGTGTGACAGCGGCTATT........................................................................................... | 24 | 1 | 1867.00 | 1867 | 0 | 1867 |
| ........................................................................TTGTGCGTGTGACAGCGGCTATT........................................................................................... | 23 | 1 | 1631.00 | 1631 | 0 | 1631 |
| ........................................................................TTGTGCGTGTGACAGCGG................................................................................................ | 18 | 1 | 1549.00 | 1549 | 0 | 1549 |
| .......................................................................CTTGTGCGTGTGACAGCG................................................................................................. | 18 | 1 | 153.00 | 153 | 0 | 153 |
| .........................................................................TGTGCGTGTGACAGCGGCTAT............................................................................................ | 21 | 1 | 49.00 | 49 | 0 | 49 |
| ..........................................................................GTGCGTGTGACAGCGGCTAT............................................................................................ | 20 | 1 | 30.00 | 30 | 0 | 30 |
| ...................................AGCTGCTGGCCACTGCACAAGA................................................................................................................................. | 22 | 1 | 29.00 | 29 | 2 | 27 |
| ...................................AGCTGCTGGCCACTGCACAAGAT................................................................................................................................ | 23 | 1 | 28.00 | 28 | 2 | 26 |
| .........................................................................TGTGCGTGTGACAGCGGCT.............................................................................................. | 19 | 1 | 23.00 | 23 | 0 | 23 |
| .........................................................................TGTGCGTGTGACAGCGGCTA............................................................................................. | 20 | 1 | 20.00 | 20 | 0 | 20 |