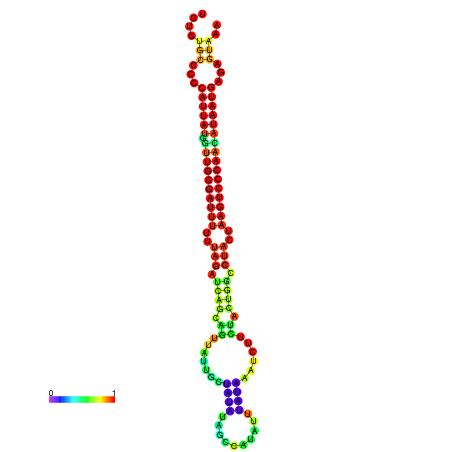
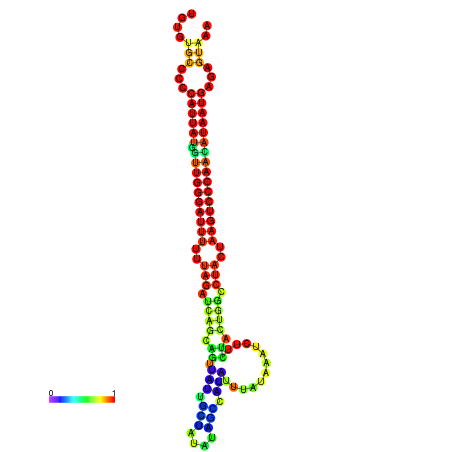
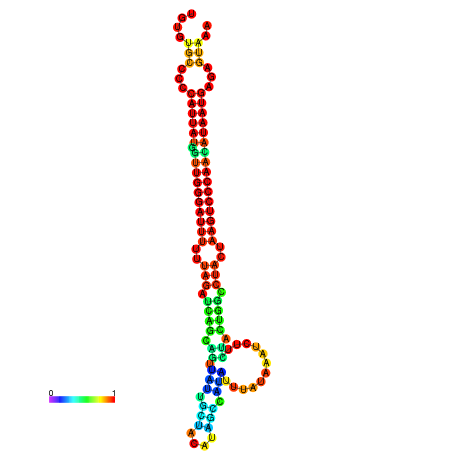
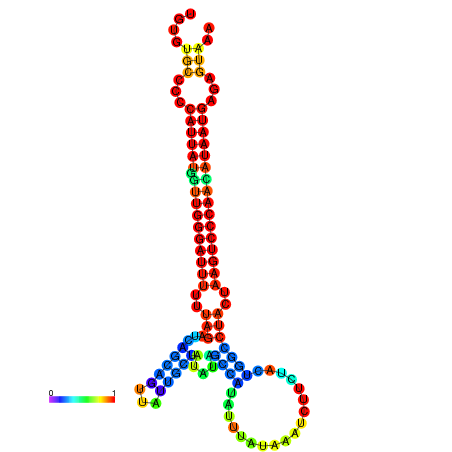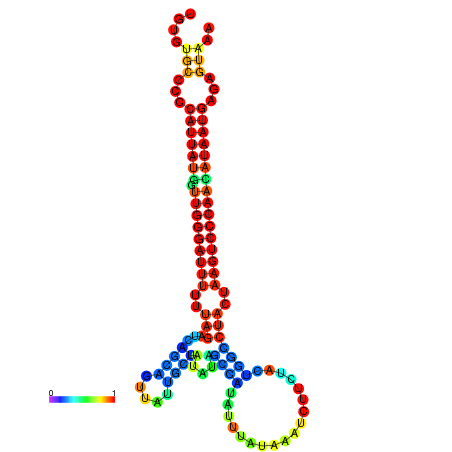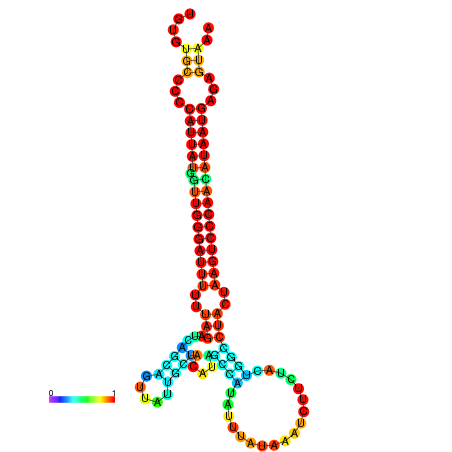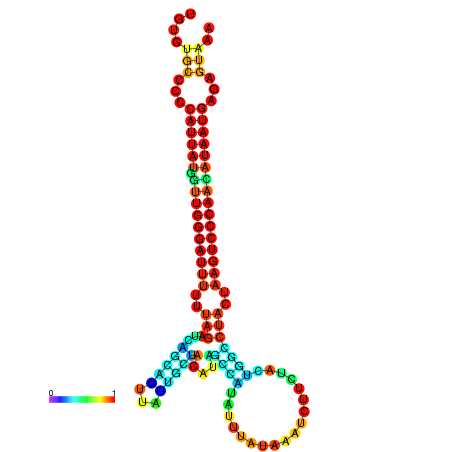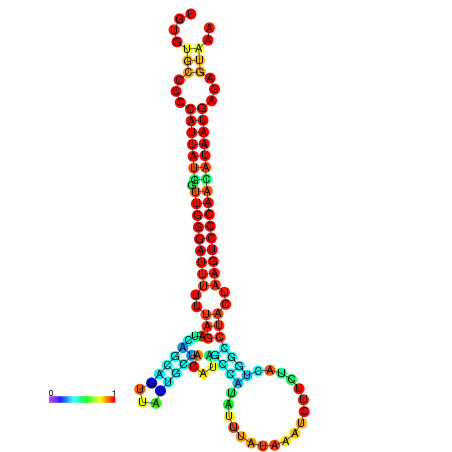| CTCAATTGGAATATTTGTGTGCACCCATTATAGTTGGGATTTTTTAGATCAGCAGTTATTGCTATACAGCCGTATTAATAAATCTTCTACTGGCCTACTAAGTCCCAACATAATGAGAGTAAAAACCACTGGCTTAACAATAA | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| .................................TTGGGATTTTTTAGATCAGC.......................................................................................... | 20 | 1 | 1074.00 | 1074 | 0 | 1074 |
| .................................TTGGGATTTTTTAGATCAGCAG........................................................................................ | 22 | 1 | 1000.00 | 1000 | 1 | 999 |
| ...............................AGTTGGGATTTTTTAGATCAGC.......................................................................................... | 22 | 1 | 754.00 | 754 | 0 | 754 |
| ...............................AGTTGGGATTTTTTAGATCAGCA......................................................................................... | 23 | 1 | 477.00 | 477 | 0 | 477 |
| ...............................AGTTGGGATTTTTTAGATCAGCAG........................................................................................ | 24 | 1 | 454.00 | 454 | 0 | 454 |
| .................................TTGGGATTTTTTAGATCAGCA......................................................................................... | 21 | 1 | 434.00 | 434 | 0 | 434 |
| ...............................AGTTGGGATTTTTTAGATCAG........................................................................................... | 21 | 1 | 381.00 | 381 | 0 | 381 |
| ................................GTTGGGATTTTTTAGATCAGC.......................................................................................... | 21 | 1 | 275.00 | 275 | 0 | 275 |
| ..................................TGGGATTTTTTAGATCAGCAGT....................................................................................... | 22 | 1 | 213.00 | 213 | 0 | 213 |
| ..................................TGGGATTTTTTAGATCAGCAG........................................................................................ | 21 | 1 | 152.00 | 152 | 0 | 152 |
| .................................TTGGGATTTTTTAGATCAGCAGT....................................................................................... | 23 | 1 | 107.00 | 107 | 0 | 107 |
| ...............................AGTTGGGATTTTTTAGATCA............................................................................................ | 20 | 1 | 65.00 | 65 | 0 | 65 |
| .......................................................................................TACTGGCCTACTAAGTCCCAAC.................................. | 22 | 1 | 44.00 | 44 | 0 | 44 |
| .................................TTGGGATTTTTTAGATCAG........................................................................................... | 19 | 1 | 38.00 | 38 | 0 | 38 |
| .......................................................................................TACTGGCCTACTAAGTCCCA.................................... | 20 | 1 | 28.00 | 28 | 0 | 28 |
| ..................................TGGGATTTTTTAGATCAGC.......................................................................................... | 19 | 1 | 24.00 | 24 | 0 | 24 |
| .......................................................................................TACTGGCCTACTAAGTCCCAA................................... | 21 | 1 | 23.00 | 23 | 0 | 23 |
| ..................................TGGGATTTTTTAGATCAGCAGTT...................................................................................... | 23 | 1 | 23.00 | 23 | 0 | 23 |
| .................................TTGGGATTTTTTAGATCAGCAGTT...................................................................................... | 24 | 1 | 19.00 | 19 | 0 | 19 |
| ................................GTTGGGATTTTTTAGATCAG........................................................................................... | 20 | 1 | 18.00 | 18 | 0 | 18 |
| ..................................TGGGATTTTTTAGATCAGCA......................................................................................... | 20 | 1 | 14.00 | 14 | 0 | 14 |
| .................................TTGGGATTTTTTAGATCA............................................................................................ | 18 | 1 | 13.00 | 13 | 0 | 13 |
| ...............................AGTTGGGATTTTTTAGATCAGCAGT....................................................................................... | 25 | 1 | 8.00 | 8 | 0 | 8 |
| ........................................................TATTGCTATACAGCCGTATTAAT................................................................ | 23 | 1 | 7.00 | 7 | 0 | 7 |
| ........................................................................................ACTGGCCTACTAAGTCCCAACAT................................ | 23 | 1 | 6.00 | 6 | 0 | 6 |
| ................................GTTGGGATTTTTTAGATCA............................................................................................ | 19 | 1 | 5.00 | 5 | 0 | 5 |
| ...............................AGTTGGGATTTTTTAGATC............................................................................................. | 19 | 1 | 4.00 | 4 | 0 | 4 |
| .......................................................................................TACTGGCCTACTAAGTCCC..................................... | 19 | 1 | 3.00 | 3 | 0 | 3 |