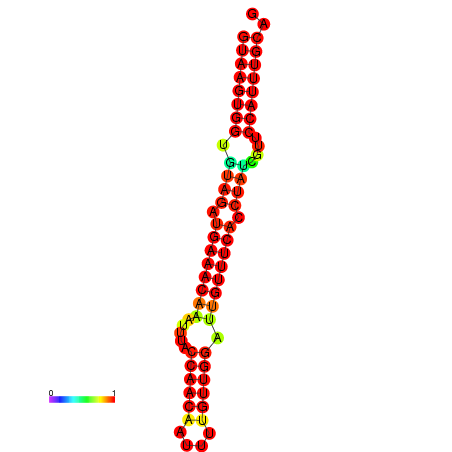
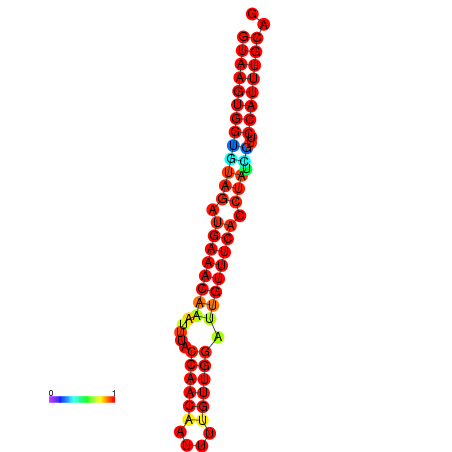
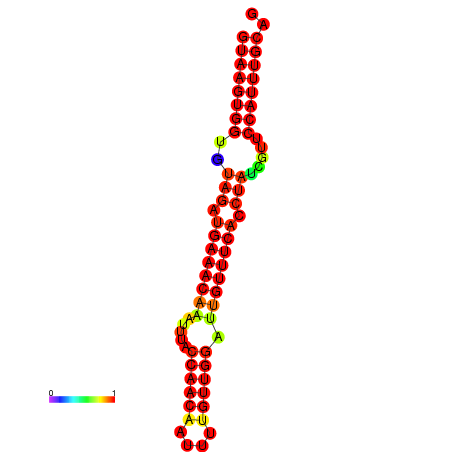
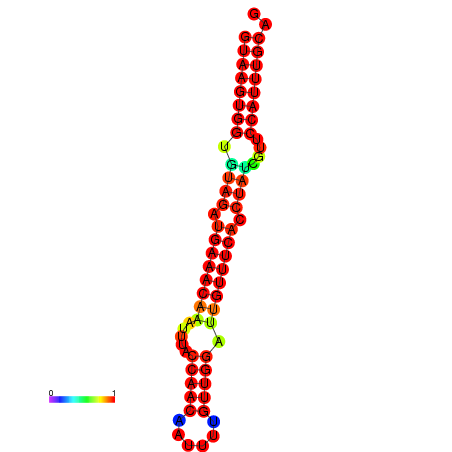
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:18118586-18118686 + | CTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAA----TTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTGGA |
| droSim1 | chr3R:17925969-17926072 + | ATTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATA-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTGGA |
| droSec1 | super_0:18476145-18476248 + | ATTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATA-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTGGA |
| droYak2 | chr3R:18962287-18962391 + | GTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATATTTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTGGA |
| droEre2 | scaffold_4820:9960766-9960869 - | CTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATA-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTCGA |
| droAna3 | scaffold_13340:9102978-9103081 + | GTTCGTGCTTAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATA-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTATGAGGACCTGGA |
| dp4 | chr2:12275938-12276041 + | GTTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTAACCAACCTTT-ATTGTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGGTACGAGGATCTCGA |
| droPer1 | super_0:8195066-8195169 - | GTTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTAACCAACCTTT-ATTGTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGGTACGAGGATCTCGA |
| droWil1 | scaffold_181130:12259759-12259863 + | ATTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATATTTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGATACGAGGATCTCGA |
| droVir3 | scaffold_13047:8363181-8363284 - | GTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTCACAACAAAT-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGATCTGGA |
| droMoj3 | scaffold_6540:27812506-27812608 + | CTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTCACAACAAT--TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGGTACGAGGATTTGGA |
| droGri2 | scaffold_15074:1256125-1256227 + | GTTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAAACTCTCAACAAT--TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGATCTGGA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:18118586-18118686 + | CTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAA----TTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTGGA |
| droSim1 | chr3R:17925969-17926072 + | ATTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATA-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTGGA |
| droSec1 | super_0:18476145-18476248 + | ATTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATA-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTGGA |
| droYak2 | chr3R:18962287-18962391 + | GTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATATTTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTGGA |
| droEre2 | scaffold_4820:9960766-9960869 - | CTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATA-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGACCTCGA |
| droAna3 | scaffold_13340:9102978-9103081 + | GTTCGTGCTTAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATA-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTATGAGGACCTGGA |
| dp4 | chr2:12275938-12276041 + | GTTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTAACCAACCTTT-ATTGTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGGTACGAGGATCTCGA |
| droPer1 | super_0:8195066-8195169 - | GTTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTAACCAACCTTT-ATTGTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGGTACGAGGATCTCGA |
| droWil1 | scaffold_181130:12259759-12259863 + | ATTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTACCAACAATATTTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGATACGAGGATCTCGA |
| droVir3 | scaffold_13047:8363181-8363284 - | GTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTCACAACAAAT-TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGATCTGGA |
| droMoj3 | scaffold_6540:27812506-27812608 + | CTTTGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAATTTCACAACAAT--TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGGTACGAGGATTTGGA |
| droGri2 | scaffold_15074:1256125-1256227 + | GTTCGTGCTCAACGGGTAAGTGGTGTAGATGAAACAAAACTCTCAACAAT--TTTTTGTTGGATTGTTTCACCTATCGTTCCATTTGCAGCTACGAGGATCTGGA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| -23.9 | -23.5 | -23.0 | -23.0 |
|---|---|---|---|
 |
 |
 |
 |
| -25.4 | -25.0 | -24.5 |
|---|---|---|
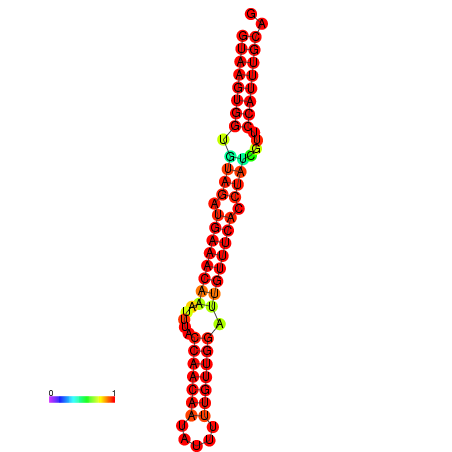 |
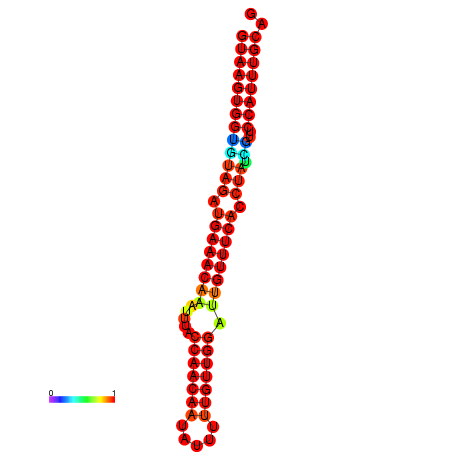 |
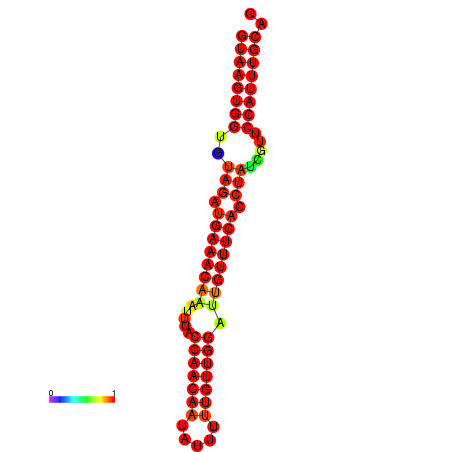 |
| -25.4 | -25.0 | -24.5 |
|---|---|---|
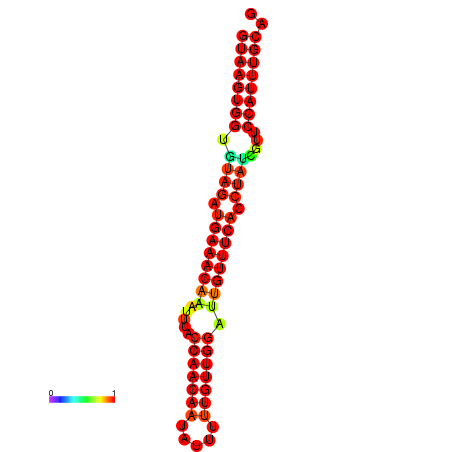 |
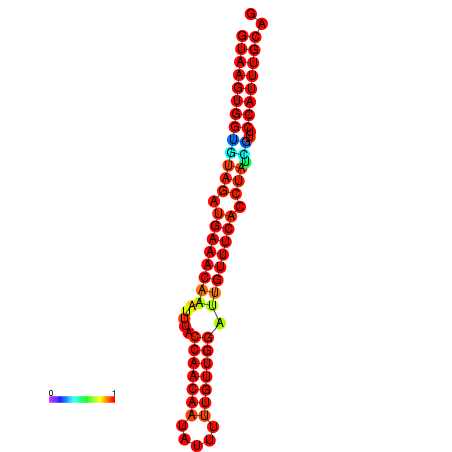 |
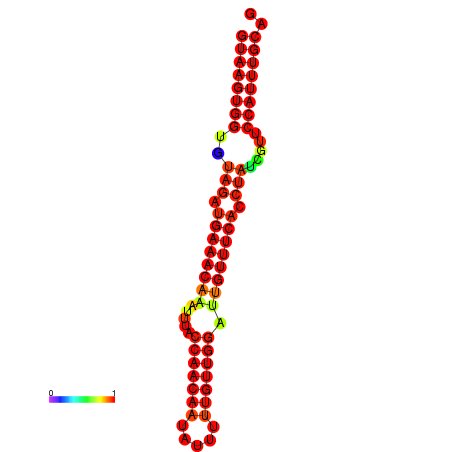 |
| -25.7 | -25.3 | -24.8 |
|---|---|---|
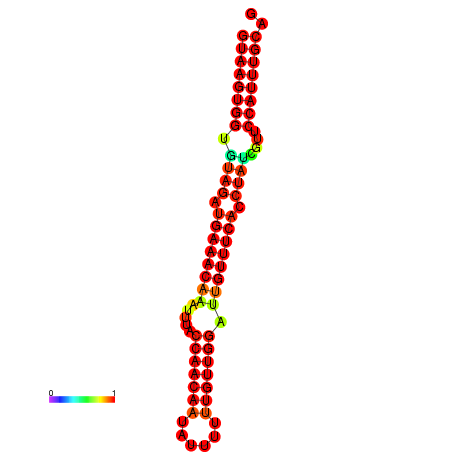 |
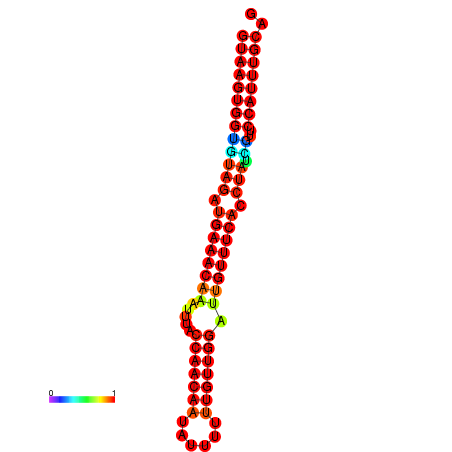 |
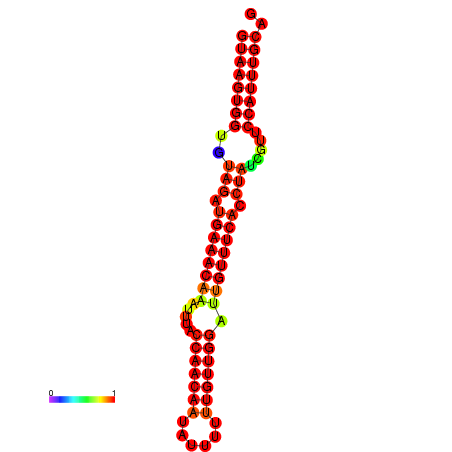 |
| -25.4 | -25.0 | -24.5 |
|---|---|---|
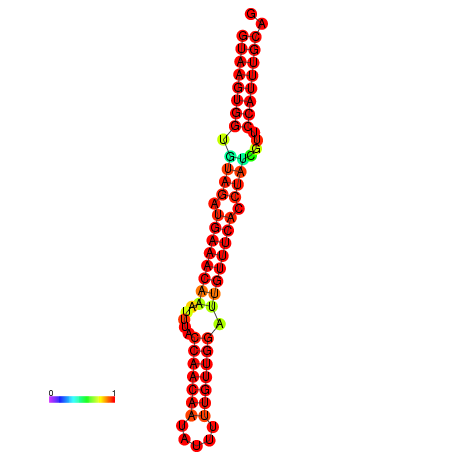 |
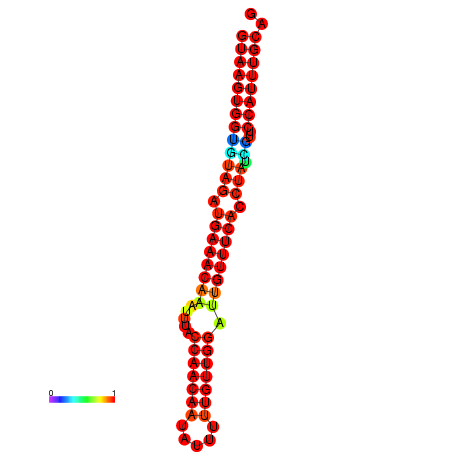 |
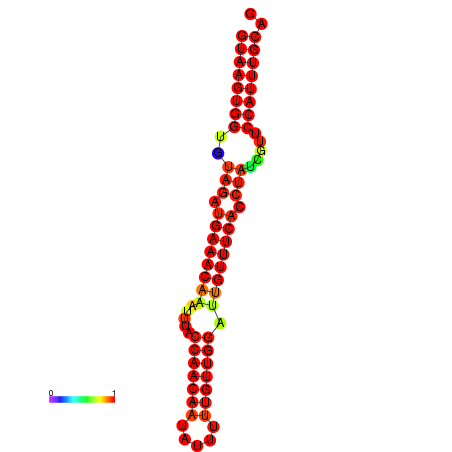 |
| -25.4 | -25.0 | -24.5 |
|---|---|---|
 |
 |
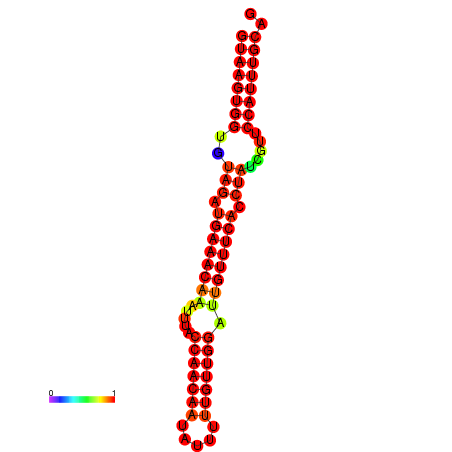 |
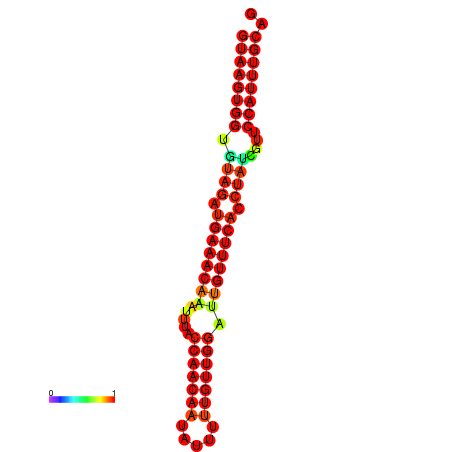
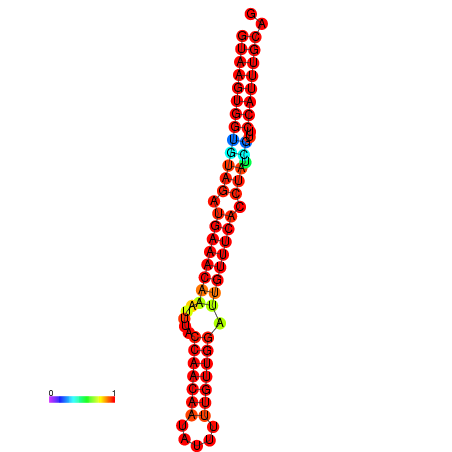
| -23.4 | -23.0 | -22.8 | -22.5 | -22.4 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
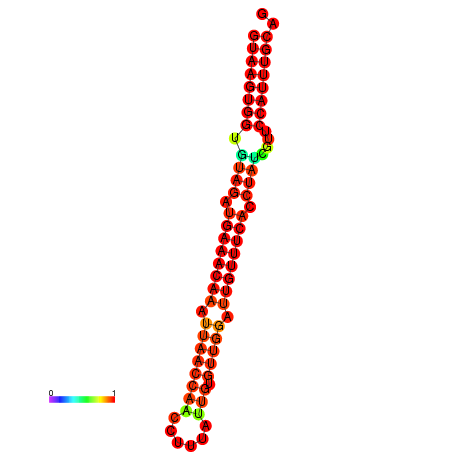
| -23.4 | -23.0 | -22.8 | -22.5 | -22.4 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| -25.7 | -25.3 | -24.8 |
|---|---|---|
 |
 |
 |
| -24.4 | -24.2 | -24.0 | -23.8 | -23.5 |
|---|---|---|---|---|
 |
 |
 |
 |
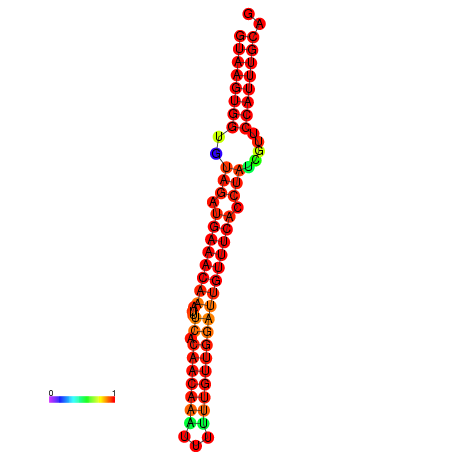 |
| -25.0 | -24.6 | -24.1 |
|---|---|---|
 |
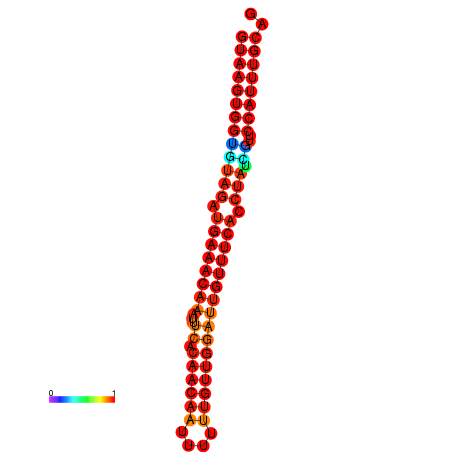 |
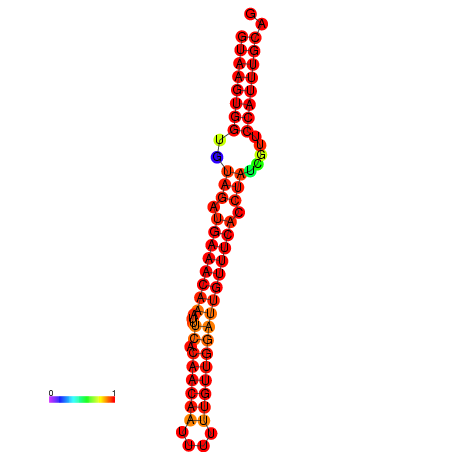 |
| -25.0 | -24.6 | -24.1 |
|---|---|---|
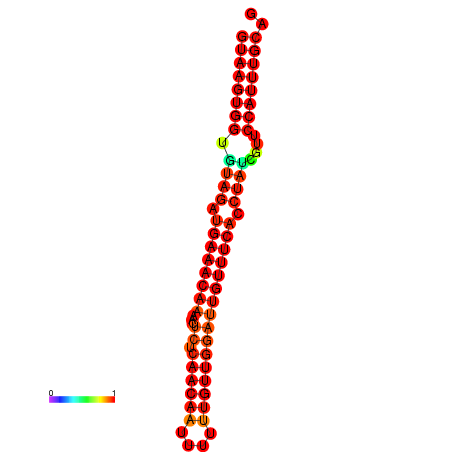 |
 |
 |
Generated: 01/05/2013 at 05:05 PM